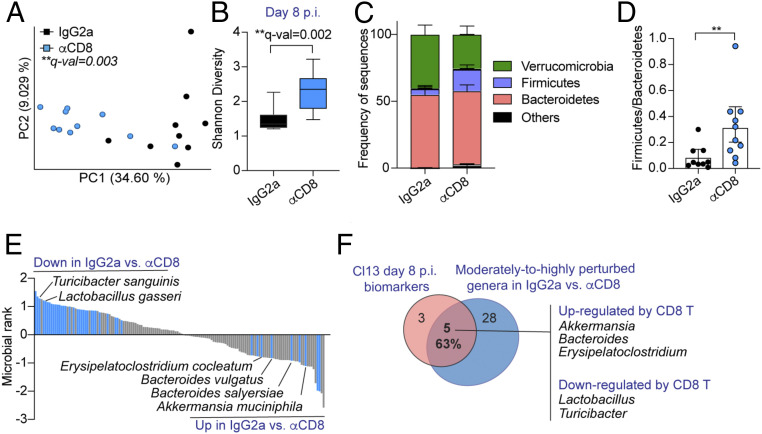
Fig. 2.
CD8 T cells drove great part of the intestinal dysbiosis identified after LCMV Cl13 infection. See also SI Appendix, Fig. S2 and Dataset S5. C57BL/6 mice were infected with LCMV Cl13, injected i.p. with isotype control (IgG2a) or CD8-depleting antibodies (αCD8) followed by shotgun metagenomics sequencing of colonic and caecal contents on day 8 p.i. (A) Beta-diversity PCoA with Jaccard distance. (B) Alpha-diversity by the Shannon diversity index. (C) Frequency of sequences at the phylum level. (D) Frequency of phylum Firmicutes over Bacteroidetes for each mouse. (E) Songbird MR analysis of species in IgG2a- vs. αCD8-treated Cl13-infected mice. Taxa with rank cutoff values of −0.5 and 0.5 that were perturbed in the same direction in two independent experimental repeats are highlighted in blue and indicated by name. (F) Overlap between Cl13-specific biomarkers at day 8 p.i. (as defined in Fig. 1F) and altered genera upon CD8 T cell depletion as defined in E. (B–D) Averages and min or max (B) or ± SEM (C and D). (A–F) Representative data from one independent experiment with n = 9 to 10 mice/group. (A) PERMANOVA with 999 permutations, (B) Kruskal–Wallis test, (D) Mann–Whitney test; *p or q-val < 0.05, **<0.01.