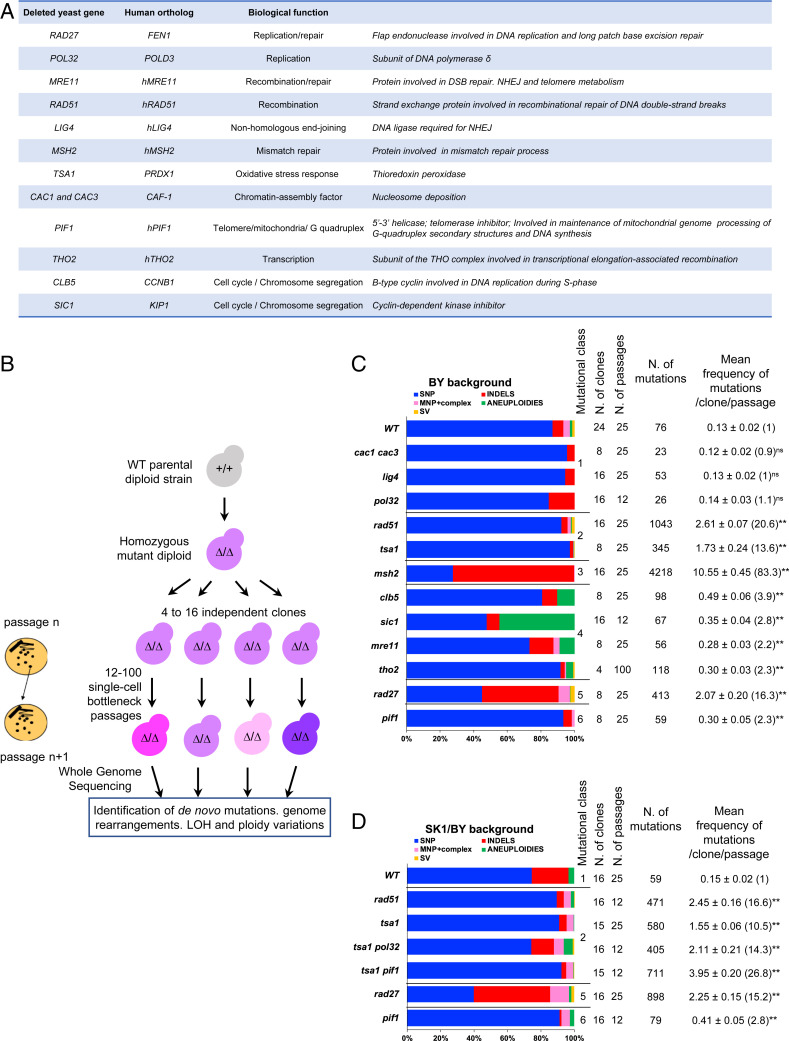
Fig. 1.
Mutational landscapes. (A) List of genes studied and their functions. (B) Experimental strategy to generate mutation accumulation lines. The WT diploid strains (BY/BY or SK1/BY background) were deleted for both copies (∆/∆) of the potential mutator gene(s). Then, 4 to 16 independent clones of the WT and ∆/∆ diploids were grown mitotically and derived for up to 100 single-cell bottleneck passages on YPD-rich medium at 30 °C (23). The genome of the resulting accumulation lines was individually sequenced by NGS and the reads were analyzed for detection of de novo mutations and genome rearrangements (SI Appendix, Materials and Methods and Fig. S1). (C and D) Mutational profiles in BY/BY and SK1/BY strains, respectively. N. of mutations: total number of de novo mutations detected in each strain, including single-nucleotide variants, small InDels, multinucleotide variants, “complex” events referring to combinations of SNPs and small InDels, chromosome aneuploidies, and structural variants (large deletions/insertions). The SNPs and small InDels comprise both heterozygous (allelic ratio ∼0.5) and apparently homozygous events (allelic ratio ∼1.0). For each mutant, the class of mutator profile and number of clones, passages, and mutations are indicated. The mean number of mutations per clone normalized to the number of passages and the SE are shown. The mutational fold variation compared with the corresponding WT is shown in parentheses. The Mann–Whitney–Wilcoxon test was performed to compare each mutant with WT (ns, not significant; **P < 0.01).