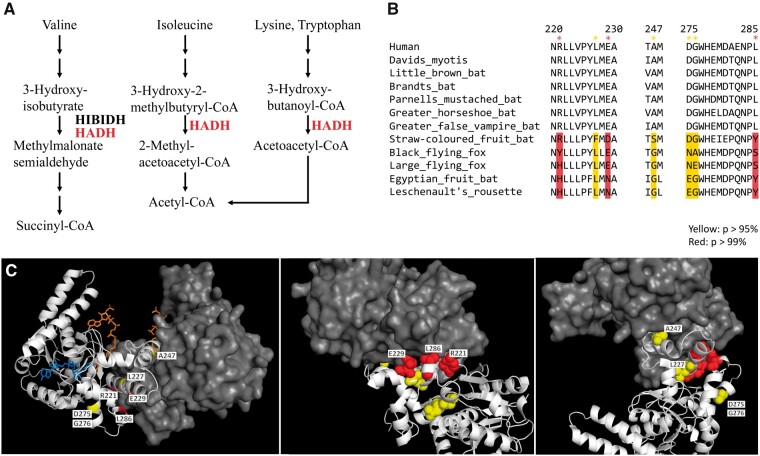
Figure 4.
Positively selected sites in HADH on megabat lineages. (A) In protein metabolism, HADH is involved in the degradation of Ile, Val, Lys, Tyr and transforms these factors into acetyl-CoA or succinyl-CoA for the TCA cycle (https://www.genome.jp/dbget-bin/www_bget?hsa: 3033). (B) The sequence alignment between the positively selected sites in HADH in the megabat lineages and microbats and human HADH. The codon alignment of all HADH sequences used in this study is available in Supplementary Alignment File S3. The sites were identified by the branch-site model on PAML. Positively selected sites are highlighted in yellow (P, >95%) and red (P-value,p >99%). (C) Positively selected residues on megabat lineages are mapped on the human HADH dimer (PDB: 1F0Y). The A chain is presented as a spherical model (yellow and red). The HADH dimer A chain is shown as a cartoon model (white) and the B chain is shown as a surface model (gray). The ligands of HADH, NAD, and acetoacetyl-CoA are shown as a stick model (blue and orange, respectively).