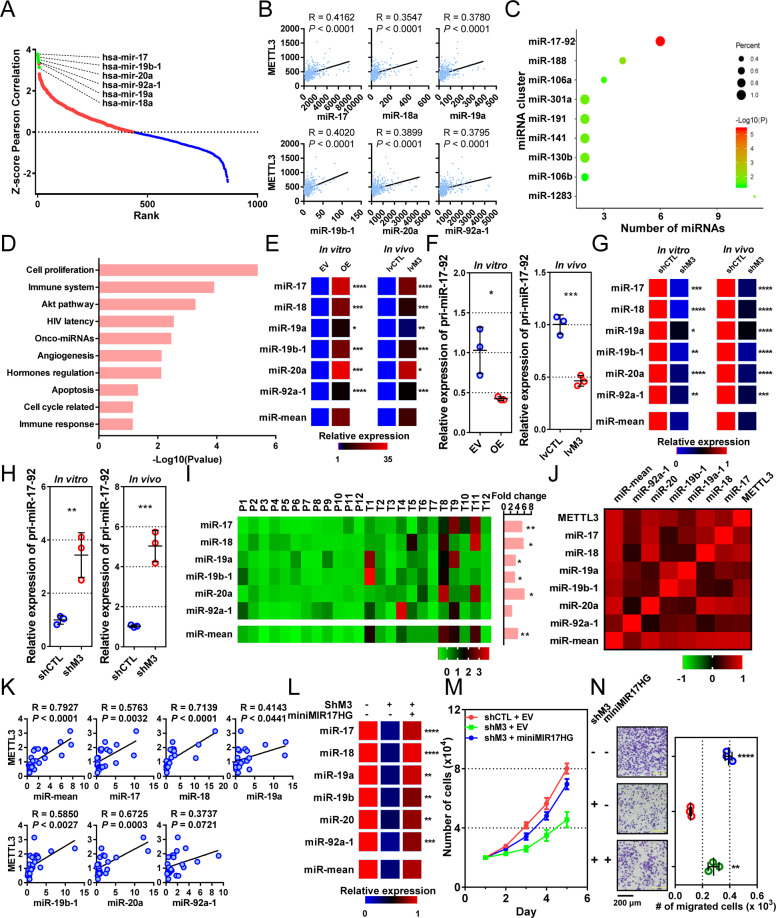
Fig. 4. METTL3 promotes tumor progression by facilitating biogenesis of miR-17-92 cluster.
a Correlation between METTL3 mRNA and all miRNAs in the TCGA-STAD database. Pearson correlation coefficients were transformed into Z-scores (y-axis) and ranked in descending order (x-axis). Blue and red dots represent negatively and positively METTL3-correlated miRNAs, respectively. Green dots represent the members of the miR-17-92 cluster. b Correlations between METTL3 (y-axis) and members of the miR-17-92 cluster (x-axis). Data were derived from the TCGA-STAD database. c MiRNA cluster enrichment results of the top METTL3-correlated miRNA (n = 76). The y-axis indicates the miRNA clusters that miRNAs were enriched in, and the x-axis indicates the numbers of METTL3-correlated miRNAs enriched in this cluster. Dot sizes indicate percentage of the METTL3-correlated miRNA in total miRNAs of the cluster, and colors indicate the P values of the enrichment. d Top 10 functional terms from METTL3-correlated miRNA enrichment. e, f Relative expressions of the miR-17-92 cluster and pri-miR-17-92 in METTL3-overexpressing (OE and lvM3) and control (EV and lvCTL) MKN-45 cells in vitro and in vivo. MiR-mean indicates mean value of the miR-17-92 cluster. g, h Relative expression of the miR-17-92 cluster and pri-miR-17-92 in METTL3-reducing (shM3) and control (shCTL) HGC-27 cells in vitro and in vivo. n = 3 for each group in e–h. i Relative expression of miR-17-92 cluster in fresh cancerous (T1–T12) and noncancerous tissues (P1–P12). Relative expression was indicated by the heatmap (left). Fold change (cancerous vs. noncancerous tissues) and statistical significance were shown on the right. j Correlation among METTL3 mRNA and miRNA miR-17-92 clusters. Correlation coefficients indicated by the heatmap. k Correlation between METTL3 mRNA (y-axis) and miR-17-92 cluster (x-axis) shown with scatter plots. l Expression of miR-17-92 cluster in HGC-27 cells expressing shM3 rescued by miniMIR17HG and empty vector (EV) transfection. m, n Proliferation and migration assays of HGC-27 cells expressing shM3 rescued by miniMIR17HG expression. The indicated significances were for comparisons between METTL3-reducing cells and those transfected with miniMIR17HG. Data are presented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.