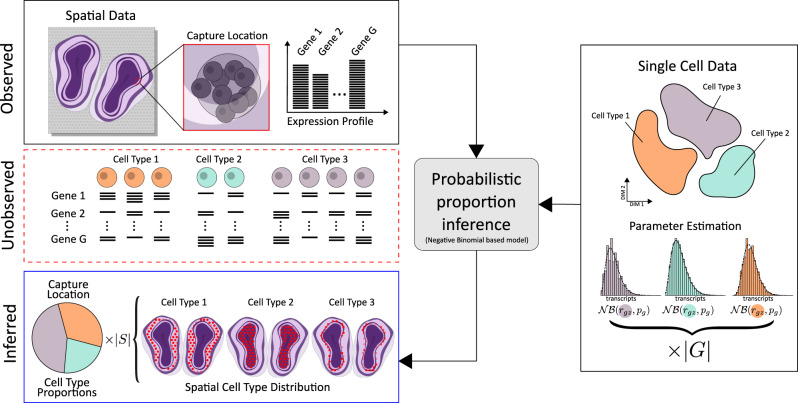
Fig. 1. The observed expression profile at each capture location is a mixture of transcripts produced by one or multiple cells, where both the number and their types are unknown.
To model the unobserved cell population at a capture location, type-specific parameters are estimated from annotated single-cell data and combined to best explain the observed data for all ∣G∣ genes. This probabilistic model, based on the negative binomial distribution, enables inference of cell type proportions at each capture location; a procedure completely free from dependence on marker genes or gene set enrichment. Doing this for all ∣S∣ capture locations, results in a map over the spatial cell type landscape of the whole tissue.