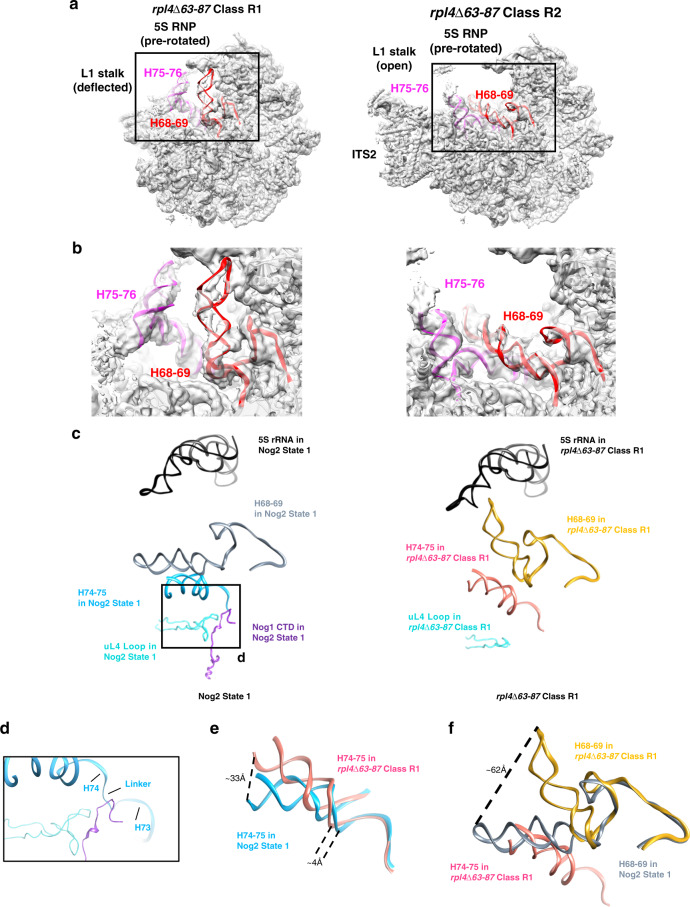
Fig. 3. Aberrant conformations of rRNA helices in rpl4∆63–87 mutant particles.
a Two major density maps (R1 and R2 particles, shown in light gray) obtained from Nog2-associated rpl4∆63–87 particles are overlaid with their respective atomic models. b Enlarged view of the conformational changes in of rRNA helices from the two major classes of the rpl4∆63–87 mutant. Compared with wild-type Nog2-particles (state 1), H75-76 (magenta) and H68-69 (red) have undergone dramatic conformational changes in class R1 but not in class R2. c rRNA helices from the atomic model of wild-type Nog2 state 1 particles are compared with those in the atomic model of the rpl4∆63–87 mutant class R1. Positions of relevant rRNA helices in relation to uL4 and Nog1 in the NPET in RPL4 wild-type (left) and rpl4∆63–87 mutant (right) Nog2 particles. Helix 74–75 (light blue for wild-type and hot pink for mutant) extend into the NPET. H74 is located in close proximity to the uL4 TD (cyan) and the CTD of Nog1 (dark purple). H74-75 are located below H68-69 (gray for wild-type and gold for mutant). The 5S rRNA (black) is shown to provide a frame of reference. d Inset showing the interaction of the TD of uL4, the Nog1 CTD, and the rRNA linker between H73 and H74. e H74-75 from rpl4∆63–87 mutant particles (hot pink) are shifted relative to those in wild-type Nog2 state 1 particles (light blue). H74 is displaced up to ~4 Å while H75 is shifted up to ~33 Å. f H75 in rpl4∆63–87 mutant particles is shifted to an aberrant position (hot pink) that clashes with the native position of H68 (gray). This displaces H68 in rpl4∆63–87 mutant particles (gold), causing it to shift up to ~62 Å.