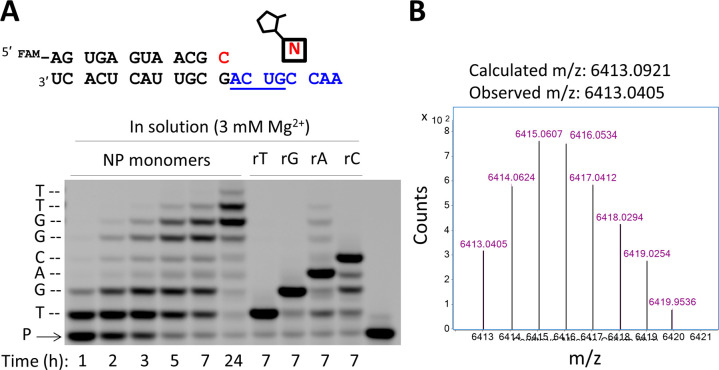
Figure 4.
Nonenzymatic primer extension on a mixed-sequence RNA template using all four 2AI activated 3′-amino nucleotides. (A) Chemical copying of an RNA template into 3′-NP-DNA. The nucleotide highlighted in red indicates the 3′-NH2-ddC at the end of the primer. The six lanes at left show the time course of the reaction. The four lanes at right show an assessment of the fidelity of 3′-NP-DNA synthesis. Each 3′-NH2-nucleotide was replaced, one at a time, with a ribonucleotide, which at 3 mM Mg2+ results in chain termination (see SI Figure S6 for full time-course). P, primer. (B) Deconvoluted monoisotopic mass distribution for the full-length primer extension product (+8) showing correct composition: calc. mass, 6413.0921 Da; obs. mass, 6413.0405 Da; error, 8.0 ppm. Primer extension reactions were carried out using 10 mM activated NP monomers (3′-NH2-2AIpddA, 3′-NH2-2AIpddG, 3′-NH2-2AIpddC, 3′-NH2-2AIpddT), 200 mM Na+-HEPES pH 8.0, 25 °C and (A) 3 mM MgCl2, (B) 50 mM MgCl2. See SI for full description of methods. The RNA templating region is indicated in blue.