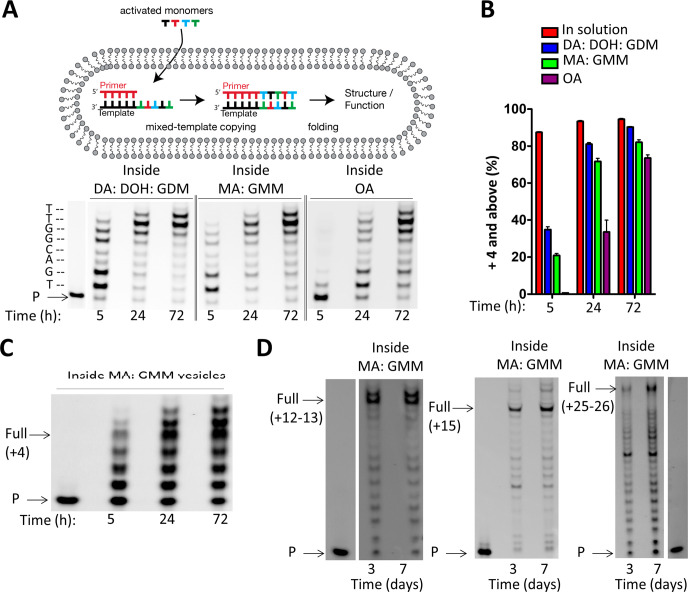
Figure 7.
Chemical copying of mixed-sequence RNA templates into 3′-NP-DNA within model protocells. (A) Schematic representation of RNA copying leading to products long enough to exhibit structure and/or function. Chemical copying of RNA template (3′-AC UGC CAA-5′) into 3′-NP-DNA within vesicles of three different membrane compositions. (B) Comparison of time course reactions in solution (SI Figure S7) and inside vesicles using 3′-NH2-2AIpddNs. Data points are reported as the mean ± s.d. from triplicate experiments. (C) Chemical copying of a randomized pool of RNA templates (Figure 3, 3′-NNNN UU-5′) into 3′-NP-DNA using 3′-NH2-2AIpddNs within MA: GMM vesicles, where N = A, G, C, or U in the template. D) Chemical copying of long mixed-sequence RNA template into 3′-NP-DNA using 3′-NH2-2AIpddNs, with templates greater than 10 (3′-ACU GAC UCC ACC G), 15 (3′-CCG CCC GAC UUC UCC GCG), or 25 (3′-ACU GAC UCC ACC GAC UGA CUC CAC CG) nucleotides in length within MA:GMM vesicles (as shown in Figure 6). It should be noted that the lanes containing the primer species only were from the same PAGE experiment as the primer extension reaction. All primer extension reactions were carried out using 10 mM activated NP monomers (3′-NH2-2AIpddA, 3′-NH2-2AIpddG, 3′-NH2-2AIpddC, 3′-NH2-2AIpddT), 50 mM MgCl2, 200 mM sodium citrate, 200 mM Na+-bicine pH 8.5, 25 °C. P, primer. Control reactions outside of vesicles are reported in the SI.