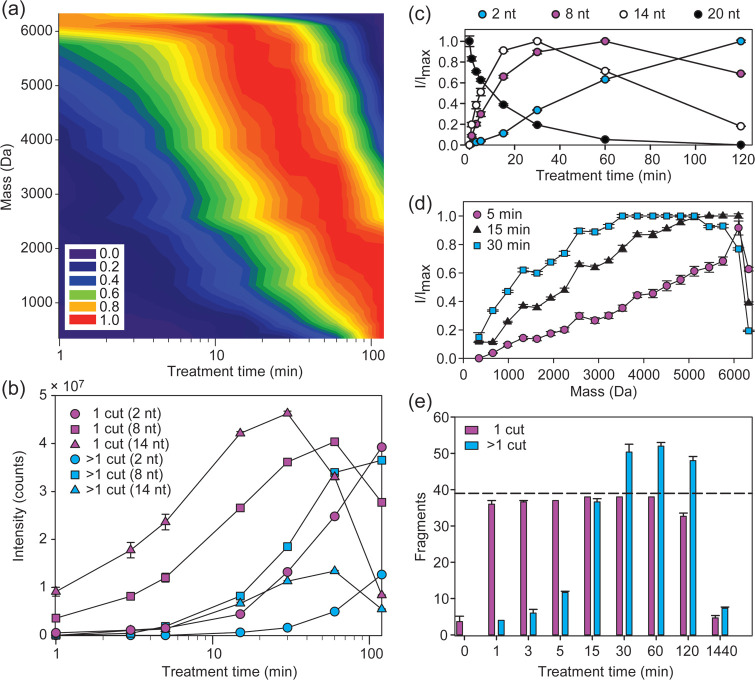
Figure 2.
Optimizing formic acid digestion to yield single-cut RNA fragments. (a) Heat map of single-cut fragment intensities in the 5′ ladder of the 20-nt RNA R1 (AUAGCCCAGUCAGUCUACGC) generated by digestion in 50% (v/v) formic acid at 40 °C. The intensities were normalized to the highest observed intensity for each fragment over a 120 min formic acid treatment. The point spacing for the time axis is shown in panel b, and the measured fragment masses set the point spacing on the mass axis. (b) Absolute intensities for selected ladder fragments arising from either one hydrolysis event (magenta) or more than one hydrolysis event (cyan) for R1. The desired single-cut fragments arise from hydrolysis of one phosphodiester bond at any position in the starting material, while undesired internal fragments arise from additional hydrolysis. (c) Normalized intensities for selected ladder fragments in the 5′ ladder over time. (d) Normalized intensities for all ladder fragments in the 5′ ladder for a 5 min (circle), 15 min (triangle) and 30 min (square) formic acid treatments. (e) Number of ladder fragments for both sequence ladders of R1 arising from one hydrolysis event (magenta) or more than one hydrolysis event (cyan) as a function of increasing formic acid treatment time. The dashed line represents the theoretical maximum number of desired fragments (39 fragments), of which we assigned 38 with stringent thresholding. The internal fragments were calculated as the number of possible fragments with unique mass. Experiments were run in triplicates, and error bars represent s.e.m.