Figure 2.
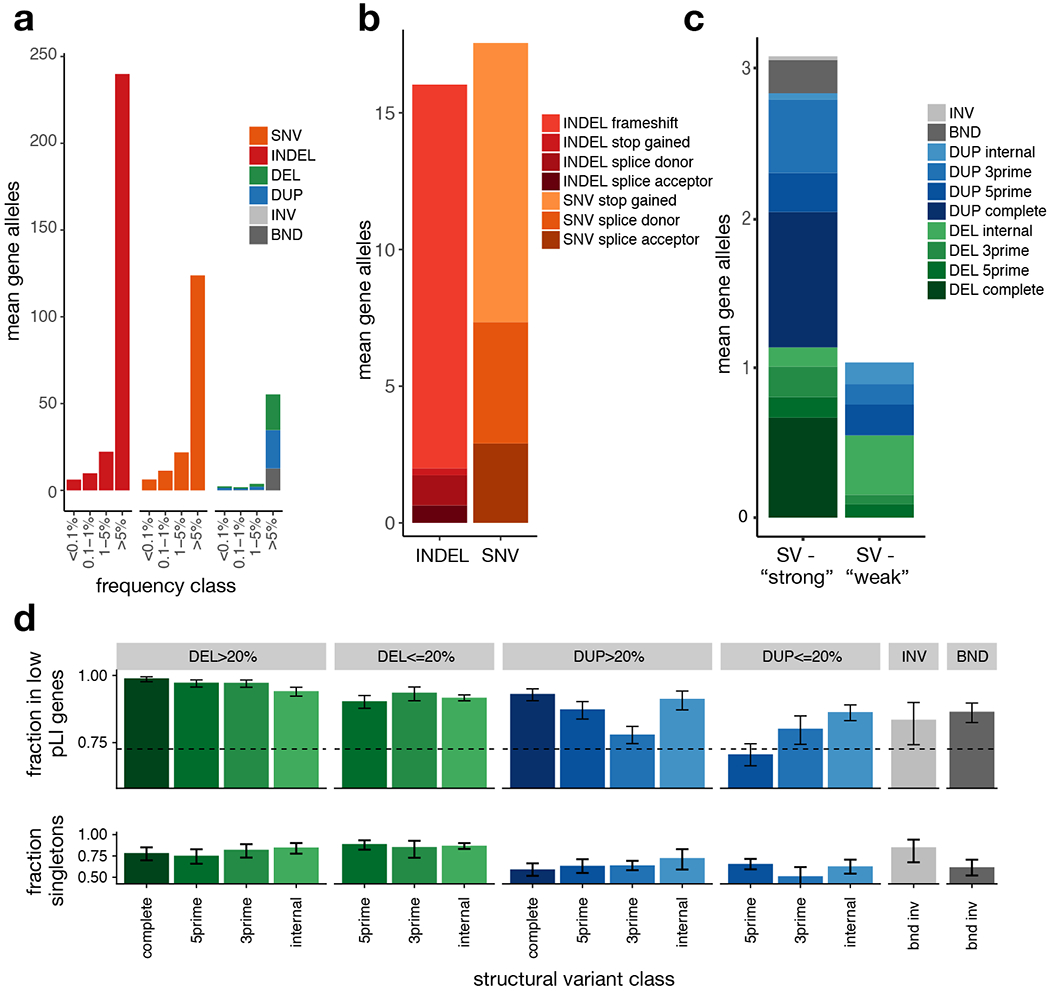
Burden of rare gene-altering SV. (a) Per-sample mean number of gene alterations by type and frequency class, in 4,298 samples. (b) Per-sample mean number of rare (<1% MAF) high-confidence PTV by type and VEP consequence. (c) Per-sample mean number of rare (<1% MAF) SV-derived gene alterations by type. DEL and DUP are classified into ‘strong’ (affecting >20% of exons of principal transcript) and ‘weak’ (affecting <20% of exons of principal transcript) and subclassified as ‘internal’ (variant overlaps at least one coding exon, but neither the 3’ nor 5’ end of the principal transcript), 3prime (variants overlaps the 3’ end of the transcript), 5prime (variant overlap the 5’ end of the transcript), and complete (variant overlaps all coding exons in principal transcript), (d) (top) Fraction of rare (<1% MAF), gene-altering variants occurring in low pLI (pLI<0.9) vs. high pLI (pLI>=0.9) genes, by type, size class, and gene region, in the B38 callset (N=14,623). Error bars indicate 95% confidence intervals (Wilson score interval). The dotted line indicates the expected fraction, assuming a uniform distribution of SV in coding exons. (bottom) Singleton rates for gene-altering variants by type in the B38 callset (N=14,623), restricted to genes with pLI>0.1. Error bars indicate 95% Wilson score confidence intervals. See Supplementary Table 5 for the number of variants in each category.
