Abstract
Purpose of Review
Dilated cardiomyopathy (DCM) frequently involves an underlying genetic etiology, but the clinical approach for genetic diagnosis and application of results in clinical practice can be complex.
Recent Findings
International sequence databases described the landscape of genetic variability across populations, which informed guidelines for the interpretation of DCM gene variants. New evidence indicates that loss-of-function mutations in filamin C (FLNC) contribute to DCM and portend high risk of ventricular arrhythmia.
Summary
A clinical framework aids in referring patients for DCM genetic testing and applying results to patient care. Results of genetic testing can change medical management, particularly in a subset of genes that increase risk for life-threatening ventricular arrhythmias, and can influence decisions for defibrillator therapy. Clinical screening and cascade genetic testing of family members should be diligently pursued to identify those at risk of developing DCM.
Keywords: Dilated cardiomyopathy, Genetic, Genetic testing, Heart failure, Mutation, Variant
Introduction
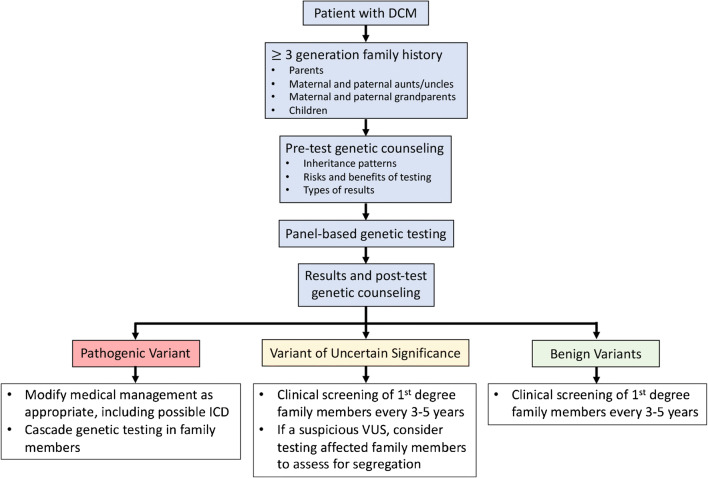
Dilated cardiomyopathy (DCM) is characterized by an enlarged ventricular cavity with reduced contractile function. By this morphological definition, DCM is often divided into “ischemic” and “nonischemic”; however, the American Heart Association and European Society of Cardiology more strictly define DCM as cardiac enlargement and dysfunction in the absence of hypertension, valvular disease, and ischemia [1, 2]. Approximately 30% of DCM was classified as idiopathic in a large survey of hospitalized patients in the USA, and a genetic cause is suspected in many cases of idiopathic and familial DCM [3]. However, the precise genetic etiology is determined in only ~ 15–35% of individuals with DCM, which illustrates the genetic heterogeneity of DCM [4, 5]. The decision to pursue genetic testing, the interpretation of results, and the appropriate clinical actions based on those results are complex. This review will address these complexities and provide a framework for applying DCM genetic testing in clinical practice (Fig. 1).
Fig. 1.
Framework for genetic testing in dilated cardiomyopathy (DCM). ICD, implantable cardioverter defibrillator; VUS, variant of uncertain significance
Genetics 101
The human genome contains 3 billion base pairs in each cell of the body, and about 3 million base pairs are different from one unrelated person to another [6]. DNA variants occur throughout the genome. However, a small percentage of variants, generally those in gene coding regions, represent the majority of variants that contribute to DCM. Single nucleotide variants that cause disease are often very rare in the population and have a large effect on phenotype. Loss-of-function (LOF) variants include the following: (1) insertions and deletions that result in a frameshift and downstream premature stop codon, (2) nucleotide substitutions that change an amino acid to a premature stop codon, and (3) canonical splice site variants that alter mRNA splicing. Missense variants lead to a non-terminating amino acid substitution within the encoded protein, with subsequent effects that range from minimal to large. Overall, missense variants that cause DCM have deleterious effects on protein structure and function. While LOF variants result in a phenotype for many genes, these results require careful assessment; for example, missense variants in MYH7 contribute to DCM, while LOF variants in MYH7 have not been observed as major drivers of DCM [7].
Chromosome pairs contain genomic DNA; in humans, 22 pairs comprise the autosomal chromosomes, while one chromosome pair (X and Y) refers to the sex chromosomes. In addition, mitochondria contain a distinct genome that encodes a subset of proteins involved in oxidative phosphorylation. Autosomal dominant and autosomal recessive inheritance patterns pertain to genes on autosomal chromosomes, while X-linked dominant and X-linked recessive inheritances refer to genes on the X chromosome. Because mitochondria are maternally inherited from the oocyte, mitochondrial inheritance occurs through the mother.
A person’s DNA sequence (genotype) and their observable traits (phenotype) are related. However, the presence of the same gene mutation may result in different phenotypes among individuals. Penetrance describes the likelihood of a particular mutation to cause disease; if not all individuals with the mutation show a disease phenotype, then the gene has reduced penetrance. Variable expressivity describes the breadth of phenotypes that are found with the same gene mutation. Several factors affect penetrance and expressivity of a particular gene mutation: secondary variants within the genome, epigenetic modulation, and environmental exposures all contribute to the phenotypic heterogeneity of DCM [8].
Brief Overview of DCM
DCM is defined as an enlarged left ventricle with reduced contractile function in the absence of hypertension, valvular disease, or ischemia [1, 2]. DCM prevalence was reported as ~ 1:2500 in an epidemiological study from 1974 to 1985 in Olmsted County, MN, USA, but more recent analyses estimate a prevalence of ~ 1:250 [8, 9]. By definition, the diagnosis of DCM requires imaging to reveal a dilated ventricle and ejection fraction of < 50%. Additional testing evaluates for ischemia, hypertension, valvular abnormalities, metabolic disease, endocrine disease, and exposure to toxins; idiopathic DCM refers to the absence of these etiologies [1]. Primary management of DCM includes beta blockers, angiotensin-converting enzyme (ACE) inhibitors, angiotensin II receptor blockers (ARBs) without or with neprolysin inhibition (ARNI), and sodium-glucose cotransporter 2 (SGLT2) inhibitors [10, 11]. Implantable cardioverter defibrillator (ICD) therapy is indicated for patients with an ejection fraction of ≤ 35% and New York Heart Association Class II–III symptoms despite optimal medical management [12].
While medical therapies have improved outcomes, the mortality rate of DCM remains high at 3.6 per 100 patient-years in men and 2.3 per 100 patient-years in women [13].
Approach for DCM Genetic Testing
Why Test?
Many factors drive the decision for genetic testing in DCM despite the current lack of gene therapies. First, genetic testing may provide a diagnosis. Second, the results of genetic testing can change medical management. For example, patients with mutations in LMNA carry a high burden of ventricular tachycardia, and ICD therapy is recommended by the European Society of Cardiology guidelines [14, 15]. Third, cascade genetic testing in family members can identify those at risk of developing cardiomyopathy. Fourth, results of genetic testing can be used for prenatal genetic counseling and preimplantation genetic diagnosis. Finally, genetic testing is cost-effective: A human whole genome can be sequenced for approximately $1000 in 2019 [16].
Who to Test?
Patients diagnosed with DCM at a young age, especially those with a family history of DCM, are at increased risk of a genetic etiology. Pathogenic (i.e., disease-causing) gene variants have been observed in 15–25% of individuals with isolated idiopathic DCM and in 20–40% of individuals with familial DCM [4, 17•, 18]. Therefore, any individual with DCM diagnosed by the age of 60 should be referred for genetic testing. Patients with a family history of DCM in a first-degree relative should undergo clinical screening with echocardiography; a family member with any ventricular abnormality should also be referred for genetic testing [19]. In keeping with this concept, isolated left ventricular dysfunction (defined as reduced ejection fraction without left ventricular dilation) has been associated with an increased burden of pathogenic variants in DCM-causing genes [20], and it is reasonable to refer individuals younger than 40 with unexplained left ventricular dysfunction.
Persistent tachycardia, alcohol, or chemotherapeutic agents are well-described causes of DCM, yet not all patients exposed to these cardiac stressors develop DCM; in some individuals, an underlying genetic variant may lower the threshold for DCM [1]. Likewise, genetic etiologies were found to contribute to a portion of peripartum cardiomyopathy (PPCM) without recovery of systolic function [21–23]. Because the presence of a DCM pathogenic variant has a large impact on management for the patient and family members, the threshold for genetic testing should be low in the clinical scenarios described above. On the other hand, the role for genetic testing remains unclear for patients with transient DCM in the setting of critical illness or isolated PPMC with recovered systolic function; in these cases, referral for genetic counseling and shared decision-making regarding genetic testing is a reasonable approach.
How are genetic testing results interpreted, and what types of results are possible?
Genetic testing does not yield a simple “positive” or “negative” result; rather, results must be interpreted in the context of population, segregation, computational, and functional data. Public databases of human genetic variation, including the Genome Aggregation Database (gnomAD; http://gnomad.broadinstutite.org) and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar), provide population data. The Genome Aggregation Database comprises over 125,000 exome sequences and over 15,000 whole genome sequences from populations around the world [24••]. ClinVar provides interpretations and supporting evidence for variants identified during genetic testing [25]. The MalaCards database (https://www.malacards.org) integrates data from 74 sources to generate rich clinical and genetic annotation for human diseases [26]. The Clinical Genome Resource (ClinGen; https://www.clinicalgenome.org/) presents expertly curated reports of clinically relevant genes and their variants; MYH7 represents the first cardiomyopathy gene with a variant analysis in ClinGen [27, 28]. In terms of segregation data, three-generation family histories provide correlations between genotype and phenotype. The American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) created an assessment scheme based on these data, which is used to classify a genetic variant as benign, likely benign, variant of uncertain significance, likely pathogenic, and pathogenic [29••].
While the details of the ACMG/AMP variant classification scheme are beyond the scope of this review, several key features can help clinicians understand the clinical implications of genetic testing results. A “negative” result on gene panel testing means that only Benign variants were observed for all the genes on that panel. Importantly, a negative genetic test does not rule out a genetic cause; the patient may have a DCM-causing mutation in a gene not yet on the panel, and family members remain at risk for genetic DCM. A pathogenic variant (mutation) is one with strong evidence that the variant causes disease. Examples of strong evidence include segregation of the genotype with phenotype in at least 5 family members across three generations, multiple independent observations of the variant associated with the phenotype in the literature, and experimental data that demonstrate loss of protein function or abnormal protein function in the presence of the variant [29••].
A variant of uncertain significance (VUS) represents a nucleotide change in a gene that is observed at a very low population frequency (less than 1 in 100,000 individuals), but the specific nucleotide change is not known to contribute to the disease phenotype. Importantly, a VUS is not the same as a benign result and should not automatically be considered absent of risk. The interpretations provided by genetic testing laboratories are performed with minimal clinical information about the patient; by contrast, the clinician is able to apply detailed knowledge of the patient’s age at diagnosis, family history, and clinical course to refine their interpretation of a VUS. A VUS might be considered “suspicious” or “trending pathogenic” if the variant is not observed in the Genome Aggregation Database and multiple family members have DCM. If the VUS segregates with a DCM phenotype in enough family members, then that VUS can be reclassified as a likely pathogenic variant [30].
Importance of Genetic Counseling
Genetic counseling should be a part of all genetic testing. Counseling involves a discussion of (1) inheritance patterns; (2) the types of results; (3) potential benefits including genetic diagnosis, changes in medical management, and family testing; and (4) risks of testing. In terms of risks, the 2008 Genetic Information Nondiscrimination Act (GINA) prohibits employers and health insurance companies from using a person’s genetic information to discriminate; however, life insurance companies are not prohibited from seeking a person’s genetic testing results to alter rates [31]. Counseling on these topics should be completed before testing is sent, and additional counseling occurs when results are returned to the patient. Specifically, if a pathogenic or likely pathogenic variant is detected, then testing for that variant should be offered to all first-degree family members. The lack of a pathogenic or likely pathogenic variant does not rule out a genetic cause, as variants in genes not known to cause DCM may be present; in this situation, the patients should inform their first-degree family members, and those family members should undergo clinical screening for DCM every three to five years. Genetic counselors represent a critical resource for providing this information to patients [32].
Genetic testing results may drive additional post-test counseling and recommendations from the cardiologist, such as consideration for ICD therapy and exercise restriction. For individuals with DCM and a pathogenic/likely pathogenic variant in a gene correlated with life-threatening arrhythmias (LMNA, FLNC, RBM20, DSP), exercise should be restricted to recreational activities [33]. By contrast, patients with DCM and a pathogenic/likely pathogenic variant in other genes may compete in sports if they are asymptomatic, ejection fraction ≥ 40%, late gadolinium enhancement ≤ 20% on MRI, and no history of unexplained syncope or significant ventricular ectopy on ambulatory ECG and exercise stress testing [33, 34]. Finally, family members identified as genotype-positive but phenotype-negative may participate in competitive sports, but they should undergo annual clinical screening [33].
Genes Implicated in Monogenic Dominant DCM
The likelihood of detecting a likely pathogenic or pathogenic variant using a cardiomyopathy gene panel had been reported at ~ 37% for DCM, although this analysis was completed before publication of large international sequence databases [4, 24••, 35]. More recently, the Dilated Cardiomyopathy Precision Medicine Study reported a likely pathogenic or pathogenic variant in 15 of 97 (15.5%) probands [5]. This relatively low testing yield reflects the complexity of the genetic architecture of DCM, and it indicates that novel genes and variants underlying DCM remain to be discovered.
A number of genes with definitive and putative contributions to DCM have been described, and these genes encode proteins that function within the sarcomere, Z disc, cytoskeleton, desmosomes, organelles, and extracellular matrix (Table 1). Genes initially attributed to other types of cardiomyopathy, such as MYBPC3 for hypertrophic cardiomyopathy (HCM) and DSP for arrhythmogenic right ventricular cardiomyopathy (ARVC), have been shown to contribute to DCM as well [19, 36, 37•].
Table 1.
Genes implicated in DCM
| Gene | Protein |
|---|---|
| Sarcomere | |
| MYH6† | α-Myosin heavy chain |
| MYH7*† | β-Myosin heavy chain |
| TPM1*† | α-Tropomyosin |
| ACTC1*† | α-Cardiac actin |
| TNNT2*† | Cardiac troponin T |
| TNNC1* | Cardiac troponin C |
| TNNI3† | Cardiac troponin I |
| MYBPC3† | Myosin-binding protein C |
| TTN*† | Titin |
| Z disk | |
| ACTN2† | α-Actinin 2 |
| BAG3*† | BCL2-associated athanogene 3 |
| CRYAB | α-B-crystallin |
| TCAP† | Titin-cap/telethonin |
| MYPN | Myopalladin |
| CSRP3† | Muscle LIM protein |
| NEXN*† | Nexilin |
| ANKRD1† | Cardiac ankyrin repeat protein |
| LDB3*† | Cypher/ZASP |
| NEBL | Nebulette |
| Cytoskeleton | |
| DES*† | Desmin |
| VCL*† | Metavinculin |
| FLNC | Filamin C |
| Desmosomes | |
| DSC2 | Desmocollin 2 |
| DSG2† | Desmoglein 2 |
| DSP*† | Desmoplakin |
| Dystrophin complex | |
| DMD*† | Dystrophin |
| DTNA | α-Dystrobrevin |
| SGCD† | δ-Sarcoglycan |
| ILK | Integrin-linked kinase |
| FKTN† | Fukutin |
| Ion channels | |
| SCN5A*† | Type V voltage-gated cardiac Na channel |
| ABCC9† | Sulfonylurea receptor 2A, component of ATP-sensitive potassium channel |
| HCN4 | Hyperpolarization-activated cyclic nucleotide-gated potassium channel 4 |
| Sarcoplasmic reticulum and cytoplasm | |
| PLN*† | Phospholamban |
| RYR2 | Ryanodine receptor 2, Ca channel |
| DOLK | Dolichol kinase |
| RAF1† | Proto-oncogene |
| Nuclear envelope | |
| LMNA*† | Lamin A/C |
| EMD | Emerin |
| TMPO† | Lamin-associated polypeptide 2 |
| SYNE1/2 | Nesprin 1/2 |
| Nucleus | |
| EYA4† | Eyes absent 4 |
| PRDM16 | PR domain containing 16 |
| TBX20 | T-box 20 |
| RBM20*† | RNA-binding protein 20 |
| GATAD1 | GATA zinc finger domain protein 1 |
| NKX2-5† | Cardiac-specific homeobox 1 |
| LRRC10 | Leucine-rich repeat containing 10 |
| Mitochondria | |
| TAZ/G4.5† | Tafazzin |
| TXNRD2 | Thioredoxin reductase 2 |
| Lysosome | |
| LAMP2† | Lysosome-associated membrane protein 2 |
| Extracellular matrix | |
| LAMA4 | Laminin 4 |
| Other | |
| CHRM2 | Cholinergic receptor muscarinic 2 |
| MIB1 | Mitogen-activated protein kinase kinase 2 |
| TTR | Transthyretin |
As the lists of genes in commercial DCM panels grow, expert curation adds context for interpretation. Independent groups have identified genes that consistently accounted for pathogenic and likely pathogenic variants for DCM: TTN, MYH7, DSP, SCN5A, LMNA, TPM1, TNNC1, TNNT2, BAG3, PLN, RBM20, LDB3, DMD, DES, ACTC1, NEXN, and VCL [5, 38–41]. FLNC was subsequently identified as an important gene for DCM and should be included in this group [5]. In these analyses, a novel or rare variant in large population sequence databases (i.e., gnomAD) provided the first level of analysis.
Ranking genes for evidence of pathogenicity significantly increased the odds ratio of a variant appearing in an individual with DCM; the highest ranked group of genes required evidence of segregation in 5 or more family members, in vitro functional studies, and heterozygous or humanized variant animal models [40]. Each of these criteria are components of the ACMG/AMP variant classification scheme, but they are not strictly required together for a variant to meet likely pathogenic or pathogenic classification [29••]. The Heart Failure Society of America and the American Heart Association each published clinical practice resources for DCM genetic testing, which recommended testing for TTN, LMNA, MYH7, TNNT2, BAG3, RBM20, TNNC1, TNNI3, TPM1, SCN5A, and PLN [17•, 18, 42•]. However, next-generation sequencing technology allows for the sequencing of dozens of genes in parallel at virtually no increased cost. Therefore, when choosing a genetic testing panel, using the largest available panel will maximize the likelihood of detecting a likely pathogenic or pathogenic variant. This approach also increases the likelihood of variants of uncertain significance, but observational studies do not detect an adverse effect on patients upon learning of VUSs [43, 44]. Furthermore, the detection of suspicious VUSs creates an opportunity for resolution of the VUS toward pathogenic or benign if other affected family members are tested. In keeping with this concept, the Heart Failure Society of America resource recommended the inclusion of HCM and ARVC genes when testing for DCM, with the acknowledgment that a larger number of VUSs will be identified [45].
Genes Implicated in DCM: Recent Updates
Titin
TTN encodes the largest protein (~ 35,000 amino acids) expressed in the body. TTN protein spans one-half of the sarcomere from the Z disc to the M line and comprises four domains: the Z disc binding region, I band domain that overlaps sarcomeric actin filaments, A band domain that overlaps myosin filaments, and the M line binding region. In 2012, the TTN gene sequence was published, and truncating variants in the A band region of TTN (TTNtv) were found to account for ~ 15–20% of all DCM [46••, 47]. At the same time, up to 3% of apparently healthy controls harbor TTNtvs, which demonstrates variable penetrance of these variants [48]. Furthermore, a recent single-center study found enrichment of TTNtvs in individuals of European ancestry with DCM, but unexpectedly these TTN truncating variants were not enriched in individuals of African ancestry in their DCM cohort [47]. Despite these complexities, TTNtvs carry prognostic significance: multiple recent studies associated TTNtvs with recovery of left ventricular systolic function and improved outcomes in the setting of guideline-directed medical therapy [49–52].
RNA-Binding Protein Motif 20 (RBM20)
RBM20 is an RNA splicing factor enriched in cardiomyocytes and skeletal muscle that regulates splicing of TTN, calcium/calmodulin dependent protein kinase II delta (CAMK2D), and ryanodine receptor 2 (RYR2); individuals with pathogenic variants in RBM20 are at high risk of DCM and ventricular arrhythmias [53–56]. Further investigation of patients from an international registry identified two regions in exons 9 and 11 in RBM20 with significant enrichment for variants associated with cardiomyopathy, ventricular and atrial arrhythmias, and sudden cardiac death [57•]. An independent study of 15 Dutch families with RBM20 pathogenic variants, all within the exon 9 and exon 11 enriched regions, found 66% penetrance of DCM and 30% with significant ventricular arrhythmia or sudden death [58]. The arrhythmogenic nature of RBM20 pathogenic variants, particularly those in exons 9 and 11, should prompt a discussion of early ICD implantation.
Desmoplakin
DSP encodes desmoplakin, a component of the desmosome that is highly expressed in the skin and cardiomyocytes. Pathogenic variants in DSP were originally described in patients with autosomal dominant ARVC, but subsequent small case series reported both missense variants and truncating variants associated with left-dominant arrhythmogenic DCM [59–65]. A large case series of 107 individuals with DSP pathogenic variants (105 with truncating variants) revealed left ventricular phenotypes in 86%, including dilation, delayed enhancement, and myocardial injury; while ventricular arrhythmias were found in 56% of individuals with DSP mutations, diagnostic criteria for ARVC were met less frequently for DSP positive as compared with PKP2-positive individuals [37•]. Myocardial injury and a distinct pattern of diffuse subepicardial delayed enhancement, even before the onset of left ventricular dilation, appear to be specific clinical findings for DSP-mediated cardiomyopathy [37•, 64, 66].
Filamin C
FLNC encodes an actin-binding intermediate filament that is highly expressed in cardiomyocytes and skeletal muscle and links membrane proteins with sarcomeres. Missense variants in FLNC have been previously associated with skeletal muscle myofibrillar myopathy and hypertrophic cardiomyopathy, but recent studies identified FLNC truncating variants (FLNCtvs) as an important driver of arrhythmogenic DCM [67••]. In the initial study of 28 probands with FLNCtvs and their genotype-positive family members, Ortiz-Genga et al. found an alarming prevalence of ventricular arrhythmias (82%) and sudden cardiac death (40 cases in 21 of 28 families) [67••]. Functional studies supported FLNC haploinsufficiency as the mechanism driving arrhythmogenic DCM [68, 69]. Multiple groups have reported FLNCtvs in probands and families with DCM and ventricular arrhythmias [68–74], which provides strong evidence for FLNCtv pathogenicity in arrhythmogenic DCM.
These recent developments in the genetic architecture of DCM underscore the importance of genetic evaluation in DCM. LMNA and SCN5A have long known to be associated with life-threatening ventricular arrhythmias, and the studies of RBM20, DSP, and FLNC discussed here clearly demonstrate significant arrhythmia and sudden death risks. In terms of clinical management, defibrillator therapy should be considered early in patients with LMNA, SCN5A, RBM20, DSP, or FLNC pathogenic variants, even before significant dilation and left ventricular systolic dysfunction have occurred.
Emerging Concepts in DCM Genetic Testing
Effects of population distributions within sequence databases on variant interpretation
The likelihood of detecting a likely pathogenic or pathogenic variant in DCM is ~ 15–25% for isolated DCM and ~ 20–40% for familial DCM, which reflects the complexity and genetic heterogeneity of DCM [4, 5, 35]. Importantly, genetic variation across populations affects variant interpretation. The vast majority of early genetic testing was performed in individuals of European non-Finnish ancestry; as worldwide testing across diverse populations has increased, many variants observed at low frequency in the European Non-Finnish population were observed in other populations at higher frequency than predicted for a rare disease. Analysis of cardiomyopathy testing at the Laboratory for Molecular Medicine revealed a lower likelihood of detecting a likely pathogenic or pathogenic variant—and a higher likelihood of a VUS—in individuals of African ancestry [75]. Furthermore, in individuals of African and Latino ancestry, VUSs in medically actionable cardiomyopathy genes are associated with clinical findings such as increased left ventricular diameter in systole and diastole [76]. As described above in a single DCM referral center, TTN truncating variants were enriched in individuals of European ancestry but not in individuals of African ancestry; reasons for this unexpected observation require further study [47]. In 2020, the population distribution in Genome Aggregation Database remained uneven: Individuals of European Non-Finnish ancestry accounted for 45% of exomes and genomes, while individuals of African, Latino, East Asian, and South Asian ancestry accounted for 9%, 13%, 7%, and 11%, respectively (the remaining data comprise Ashkenazi Jewish, Finnish, and “other” ancestries). Accurate interpretation of variants depends upon representative sampling of a sufficient number of individuals across multiple populations; inclusion of more non-European individuals and families remains an ongoing goal within the field [5]. Expansion of genetic testing in non-European individuals and families will improve classification, since many variants meet likely pathogenic or pathogenic classification by combining evidence from multiple families.
Polygenic Risk Scores
Given the genetic heterogeneity of DCM, it is reasonable to predict that common variants with minor allele frequency of > 1% will partially contribute to DCM phenotypes. Genome-wide association studies (GWAS) have identified several common single nucleotide variants associated with idiopathic DCM at the population level [77–79].
Polygenic risk scores (PRSs) calculate a relative risk of developing disease based on an individual’s number of common variants associated with that disease [80]. Currently, the relatively small number of common variants associated with DCM limits the applications of PRSs, but this application will likely augment DCM risk prediction as additional GWAS are completed.
Population Databases and Refining Estimates of Penetrance
As population sequence databases have increased in size, efforts have been made to use this information to refine penetrance estimates. This concept holds particular relevance in DCM, where incomplete and age-dependent penetrance remain important concepts in counseling patients who are genotype-positive but phenotype-negative. To date, methods to use population-based whole exome and whole genome sequencing for penetrance estimates show variability [81–83]. The Genome Aggregation Database does not contain phenotypic data, but other programs including NHLBI Trans-Omics for Precision Medicine (TOPMed), NIH All of Us, NHGRI Genome Sequencing Project, and Harvard Medical School Genomes2People have been designed to include whole exome or whole genome sequencing with deep phenotyping information. The generation of large datasets linking genotype and phenotype from diverse populations should significantly improve estimates of penetrance.
Whole-Exome Sequencing and Whole-Genome Sequencing as Initial Genetic Testing Strategies
The most common genetic testing approach utilizes panel-based sequencing. Many genetic testing laboratories offer WES as an alternative approach [84]. However, whole-exome sequencing (WES) may not provide sufficient coverage for all genes; TNNI3 and PLN were reported to have insufficient coverage for complete variant interpretation [85]. By contrast, results from whole-genome sequencing (WGS) correlated well with panel-based sequencing, and WGS performed better than WES: WES covered only 69% of panel sequence targets, most likely due to capture bias during sample preparation and use of predefined target regions that may miss isoforms [86]. Because WES and WGS are not limited to known DCM genes, the provider must be prepared for clinically actionable likely pathogenic and pathogenic variants in non-cardiomyopathy genes. Overall, the practical applications of WES and WGS are still being developed for clinical DCM genetic testing. In the research setting, WGS remains a powerful technique for new DCM gene discovery.
Conclusion
DCM is a genetically heterogeneous condition, and the interpretation of results can be complex. Despite these complexities, results from genetic testing can have a profound impact on patient management, particularly for the arrhythmogenic subset of DCM. Furthermore, cascade genetic testing can identify family members at risk for developing DCM. Finally, expansion of testing in diverse populations will improve variant interpretation in the future.
Compliance with Ethical Standards
Conflict of Interest
The author declares that there is no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
This article is part of the Topical Collection on Cardiovascular Genomics
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
- 1.Bozkurt B, Colvin M, Cook J, Cooper LT, Deswal A, Fonarow GC, Francis GS, Lenihan D, Lewis EF, McNamara D, Pahl E, Vasan RS, Ramasubbu K, Rasmusson K, Towbin JA, Yancy C, American Heart Association Committee on Heart Failure and Transplantation of the Council on Clinical Cardiology; Council on Cardiovascular Disease in the Young; Council on Cardiovascular and Stroke Nursing; Council on Epidemiology and Prevention; and Council on Quality of Care and Outcomes Research Current diagnostic and treatment strategies for specific dilated cardiomyopathies: a scientific statement from the American Heart Association. Circulation. 2016;134(23):e579–e646. doi: 10.1161/CIR.0000000000000455. [DOI] [PubMed] [Google Scholar]
- 2.Pinto YM, Elliott PM, Arbustini E, Adler Y, Anastasakis A, Bohm M, et al. Proposal for a revised definition of dilated cardiomyopathy, hypokinetic non- dilated cardiomyopathy, and its implications for clinical practice: a position statement of the ESC working group on myocardial and pericardial diseases. Eur Heart J. 2016;37(23):1850–1858. doi: 10.1093/eurheartj/ehv727. [DOI] [PubMed] [Google Scholar]
- 3.Shore S, Grau-Sepulveda MV, Bhatt DL, Heidenreich PA, Eapen ZJ, Hernandez AF, Yancy CW, Fonarow GC. Characteristics, treatments, and outcomes of hospitalized heart failure patients stratified by etiologies of cardiomyopathy. JACC Heart Fail. 2015;3(11):906–916. doi: 10.1016/j.jchf.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Pugh TJ, Kelly MA, Gowrisankar S, Hynes E, Seidman MA, Baxter SM, Bowser M, Harrison B, Aaron D, Mahanta LM, Lakdawala NK, McDermott G, White ET, Rehm HL, Lebo M, Funke BH. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet Med. 2014;16(8):601–608. doi: 10.1038/gim.2013.204. [DOI] [PubMed] [Google Scholar]
- 5.Morales A, Kinnamon DD, Jordan E, Platt J, Vatta M, Dorschner MO, Starkey CA, Mead JO, Ai T, Burke W, Gastier-Foster J, Jarvik GP, Rehm HL, Nickerson DA, Hershberger RE, on behalf of the DCM Precision Medicine study of the DCM Consortium. Bowen DJ, Haas G, Abraham WT, Binkley PF, Hasan A, Host J, Lampert B, Smith S, Huggins GS, DeNofrio DD, Kiernan M, Fishbein D, Cheng R, Dardas T, Levy W, Mahr C, Masri S, Stempien-Otero A, Gottlieb (Ste Pl) SS, Wheeler M, Ashley E, Hofmeyer M, Tang WHW, Starling R, Owens A, Marguilies KB, Cappola T, Goldberg LR, McLean R, Moore CK, Long RC, Jimenez Carcamo J, Trachtenberg B, Ashrith G, Bhimarahj A, Sweitzer NK, Shah P, Lowes B, Stoller D, Smart F, Morris AA, Wilcox J, Pan S, Ewald GA, Aaronson KD, Wang JJ, Pamboukian S, Judge DP, Kransdorf EP, Garg S, Desvigne- Nickens P, Troendle J, Fu YP, Hindorff L. Variant interpretation for dilated cardiomyopathy: refinement of the American College of Medical Genetics and Genomics/ClinGen guidelines for the DCM precision medicine study. Circ Genom Precis Med. 2020;13(2):e002480. doi: 10.1161/CIRCGEN.119.002480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd SD. Diagnostic applications of high-throughput DNA sequencing. Annu Rev Pathol. 2013;8:381–410. doi: 10.1146/annurev-pathol-020712-164026. [DOI] [PubMed] [Google Scholar]
- 7.Walsh R, Thomson KL, Ware JS, Funke BH, Woodley J, McGuire KJ, et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2017;19(2):192–203. doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol. 2013;10(9):531–547. doi: 10.1038/nrcardio.2013.105. [DOI] [PubMed] [Google Scholar]
- 9.Codd MB, Sugrue DD, Gersh BJ, Melton LJ., 3rd Epidemiology of idiopathic dilated and hypertrophic cardiomyopathy. A population-based study in Olmsted County, Minnesota, 1975-1984. Circulation. 1989;80(3):564–572. doi: 10.1161/01.cir.80.3.564. [DOI] [PubMed] [Google Scholar]
- 10.Yancy CW, Jessup M, Bozkurt B, Butler J, Casey DE, Jr, Colvin MM, Drazner MH, Filippatos GS, Fonarow GC, Givertz MM, Hollenberg SM, Lindenfeld JA, Masoudi FA, McBride PE, Peterson PN, Stevenson LW, Westlake C. 2017 ACC/AHA/HFSA focused update of the 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology/American Heart Association task force on clinical practice guidelines and the Heart Failure Society of America. Circulation. 2017;136(6):e137–ee61. doi: 10.1161/CIR.0000000000000509. [DOI] [PubMed] [Google Scholar]
- 11.McMurray JJV, Solomon SD, Inzucchi SE, Kober L, Kosiborod MN, Martinez FA, et al. Dapagliflozin in patients with heart failure and reduced ejection fraction. N Engl J Med. 2019;381(21):1995–2008. doi: 10.1056/NEJMoa1911303. [DOI] [PubMed] [Google Scholar]
- 12.Epstein AE, DiMarco JP, Ellenbogen KA, Estes NA, 3rd, Freedman RA, Gettes LS, et al. 2012 ACCF/AHA/HRS focused update incorporated into the ACCF/AHA/HRS 2008 guidelines for device-based therapy of cardiac rhythm abnormalities: a report of the American College of Cardiology Foundation/American Heart Association task force on practice guidelines and the Heart Rhythm Society. J Am Coll Cardiol. 2013;61(3):e6–75. doi: 10.1016/j.jacc.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Halliday BP, Gulati A, Ali A, Newsome S, Lota A, Tayal U, Vassiliou VS, Arzanauskaite M, Izgi C, Krishnathasan K, Singhal A, Chiew K, Gregson J, Frenneaux MP, Cook SA, Pennell DJ, Collins P, Cleland JGF, Prasad SK. Sex- and age- based differences in the natural history and outcome of dilated cardiomyopathy. Eur J Heart Fail. 2018;20(10):1392–1400. doi: 10.1002/ejhf.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sidhu K, Han L, Picard KCI, Tedrow UB, Lakdawala NK. Ventricular tachycardia in cardiolaminopathy: characteristics and considerations for device programming. Heart Rhythm. 2020. 10.1016/j.hrthm.2020.05.023. [DOI] [PubMed]
- 15.Priori SG, Blomstrom-Lundqvist C, Mazzanti A, Blom N, Borggrefe M, Camm J, et al. 2015 ESC Guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: the task force for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death of the European Society of Cardiology (ESC). Endorsed by: Association for European Paediatric and Congenital Cardiology (AEPC) Eur Heart J. 2015;36(41):2793–2867. doi: 10.1093/eurheartj/ehv316. [DOI] [PubMed] [Google Scholar]
- 16.Wetterstrand KA. DNA sequencing costs: data from the NHGRI genome sequencing program (GSP) www.genome.gov/sequencingcostsdata. Accessed August 5, 2020.
- 17.Hershberger RE, Givertz MM, Ho CY, Judge DP, Kantor PF, McBride KL, et al. Genetic evaluation of cardiomyopathy: a clinical practice resource of the American College of Medical Genetics and Genomics (ACMG) Genet Med. 2018;20(9):899–909. doi: 10.1038/s41436-018-0039-z. [DOI] [PubMed] [Google Scholar]
- 18.Hershberger RE, Givertz MM, Ho CY, Judge DP, Kantor PF, McBride KL, et al. Genetic evaluation of cardiomyopathy-a Heart Failure Society of America practice guideline. J Card Fail. 2018;24(5):281–302. doi: 10.1016/j.cardfail.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoedemaekers YM, Caliskan K, Michels M, Frohn-Mulder I, van der Smagt JJ, Phefferkorn JE, Wessels MW, ten Cate FJ, Sijbrands EJG, Dooijes D, Majoor-Krakauer DF. The importance of genetic counseling, DNA diagnostics, and cardiologic family screening in left ventricular noncompaction cardiomyopathy. Circ Cardiovasc Genet. 2010;3(3):232–239. doi: 10.1161/CIRCGENETICS.109.903898. [DOI] [PubMed] [Google Scholar]
- 20.Hazebroek MR, Krapels I, Verdonschot J, van den Wijngaard A, Vanhoutte E, Hoos M, Snijders L, van Montfort L, Witjens M, Dennert R, Crijns HJGM, Brunner-la Rocca HP, Brunner HG, Heymans S. Prevalence of pathogenic gene mutations and prognosis do not differ in isolated left ventricular dysfunction compared with dilated cardiomyopathy. Circ Heart Fail. 2018;11(3):e004682. doi: 10.1161/CIRCHEARTFAILURE.117.004682. [DOI] [PubMed] [Google Scholar]
- 21.van Spaendonck-Zwarts KY, van Tintelen JP, van Veldhuisen DJ, van der Werf R, Jongbloed JD, Paulus WJ, et al. Peripartum cardiomyopathy as a part of familial dilated cardiomyopathy. Circulation. 2010;121(20):2169–2175. doi: 10.1161/CIRCULATIONAHA.109.929646. [DOI] [PubMed] [Google Scholar]
- 22.Morales A, Painter T, Li R, Siegfried JD, Li D, Norton N, Hershberger RE. Rare variant mutations in pregnancy-associated or peripartum cardiomyopathy. Circulation. 2010;121(20):2176–2182. doi: 10.1161/CIRCULATIONAHA.109.931220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krul SP, van der Smagt JJ, van den Berg MP, Sollie KM, Pieper PG, van Spaendonck-Zwarts KY. Systematic review of pregnancy in women with inherited cardiomyopathies. Eur J Heart Fail. 2011;13(6):584–594. doi: 10.1093/eurjhf/hfr040. [DOI] [PubMed] [Google Scholar]
- 24.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Jang W, Karapetyan K, Katz K, Liu C, Maddipatla Z, Malheiro A, McDaniel K, Ovetsky M, Riley G, Zhou G, Holmes JB, Kattman BL, Maglott DR. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46(D1):D1062–D10D7. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rappaport N, Twik M, Plaschkes I, Nudel R, Iny Stein T, Levitt J, Gershoni M, Morrey CP, Safran M, Lancet D. MalaCards: an amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 2017;45(D1):D877–DD87. doi: 10.1093/nar/gkw1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rehm HL, Berg JS, Brooks LD, Bustamante CD, Evans JP, Landrum MJ, Ledbetter DH, Maglott DR, Martin CL, Nussbaum RL, Plon SE, Ramos EM, Sherry ST, Watson MS, ClinGen ClinGen--the clinical genome resource. N Engl J Med. 2015;372(23):2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelly MA, Caleshu C, Morales A, Buchan J, Wolf Z, Harrison SM, et al. Adaptation and validation of the ACMG/AMP variant classification framework for MYH7-associated inherited cardiomyopathies: recommendations by ClinGen's inherited cardiomyopathy expert panel. Genet Med. 2018;20(3):351–359. doi: 10.1038/gim.2017.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cuenca S, Ruiz-Cano MJ, Gimeno-Blanes JR, Jurado A, Salas C, Gomez-Diaz I, Padron-Barthe L, Grillo JJ, Vilches C, Segovia J, Pascual-Figal D, Lara-Pezzi E, Monserrat L, Alonso-Pulpon L, Garcia-Pavia P, Inherited Cardiac Diseases Program of the Spanish Cardiovascular Research Network (Red Investigación Cardiovascular) Genetic basis of familial dilated cardiomyopathy patients undergoing heart transplantation. J Heart Lung Transplant. 2016;35(5):625–635. doi: 10.1016/j.healun.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 31.Prince AE, Roche MI. Genetic information, non-discrimination, and privacy protections in genetic counseling practice. J Genet Couns. 2014;23(6):891–902. doi: 10.1007/s10897-014-9743-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nieuwhof K, Birnie E, van den Berg MP, de Boer RA, van Haelst PL, van Tintelen JP, van Langen IM. Follow-up care by a genetic counsellor for relatives at risk for cardiomyopathies is cost-saving and well-appreciated: a randomised comparison. Eur J Hum Genet. 2017;25(2):169–175. doi: 10.1038/ejhg.2016.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelliccia A, Solberg EE, Papadakis M, Adami PE, Biffi A, Caselli S, la Gerche A, Niebauer J, Pressler A, Schmied CM, Serratosa L, Halle M, van Buuren F, Borjesson M, Carrè F, Panhuyzen-Goedkoop NM, Heidbuchel H, Olivotto I, Corrado D, Sinagra G, Sharma S. Recommendations for participation in competitive and leisure time sport in athletes with cardiomyopathies, myocarditis, and pericarditis: position statement of the sport cardiology section of the European Association of Preventive Cardiology (EAPC) Eur Heart J. 2019;40(1):19–33. doi: 10.1093/eurheartj/ehy730. [DOI] [PubMed] [Google Scholar]
- 34.Maron BJ, Udelson JE, Bonow RO, Nishimura RA, Ackerman MJ, Estes NA, 3rd, et al. Eligibility and disqualification recommendations for competitive athletes with cardiovascular abnormalities: task force 3: hypertrophic cardiomyopathy, arrhythmogenic right ventricular cardiomyopathy and other cardiomyopathies, and myocarditis: a scientific statement from the American Heart Association and American College of Cardiology. Circulation. 2015;132(22):e273–e280. doi: 10.1161/CIR.0000000000000239. [DOI] [PubMed] [Google Scholar]
- 35.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoedemaekers YM, Caliskan K, Majoor-Krakauer D, van de Laar I, Michels M, Witsenburg M, ten Cate FJ, Simoons ML, Dooijes D. Cardiac beta-myosin heavy chain defects in two families with non-compaction cardiomyopathy: linking non-compaction to hypertrophic, restrictive, and dilated cardiomyopathies. Eur Heart J. 2007;28(22):2732–2737. doi: 10.1093/eurheartj/ehm429. [DOI] [PubMed] [Google Scholar]
- 37.Smith ED, Lakdawala NK, Papoutsidakis N, Aubert G, Mazzanti A, McCanta AC, et al. Desmoplakin cardiomyopathy, a fibrotic and inflammatory form of cardiomyopathy distinct from typical dilated or arrhythmogenic right ventricular cardiomyopathy. Circulation. 2020;141(23):1872–1884. doi: 10.1161/CIRCULATIONAHA.119.044934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haas J, Frese KS, Peil B, Kloos W, Keller A, Nietsch R, et al. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur Heart J. 2015;36(18):1123–135a. doi: 10.1093/eurheartj/ehu301. [DOI] [PubMed] [Google Scholar]
- 39.Kayvanpour E, Sedaghat-Hamedani F, Amr A, Lai A, Haas J, Holzer DB, Frese KS, Keller A, Jensen K, Katus HA, Meder B. Genotype-phenotype associations in dilated cardiomyopathy: meta-analysis on more than 8000 individuals. Clin Res Cardiol. 2017;106(2):127–139. doi: 10.1007/s00392-016-1033-6. [DOI] [PubMed] [Google Scholar]
- 40.Horvat C, Johnson R, Lam L, Munro J, Mazzarotto F, Roberts AM, Herman DS, Parfenov M, Haghighi A, McDonough B, DePalma SR, Keogh AM, Macdonald PS, Hayward CS, Roberts A, Barton PJR, Felkin LE, Giannoulatou E, Cook SA, Seidman JG, Seidman CE, Fatkin D. A gene- centric strategy for identifying disease-causing rare variants in dilated cardiomyopathy. Genet Med. 2019;21(1):133–143. doi: 10.1038/s41436-018-0036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazzarotto F, Tayal U, Buchan RJ, Midwinter W, Wilk A, Whiffin N, Govind R, Mazaika E, de Marvao A, Dawes TJW, Felkin LE, Ahmad M, Theotokis PI, Edwards E, Ing AY, Thomson KL, Chan LLH, Sim D, Baksi AJ, Pantazis A, Roberts AM, Watkins H, Funke B, O’Regan DP, Olivotto I, Barton PJR, Prasad SK, Cook SA, Ware JS, Walsh R. Reevaluating the genetic contribution of monogenic dilated cardiomyopathy. Circulation. 2020;141(5):387–398. doi: 10.1161/CIRCULATIONAHA.119.037661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.• Musunuru K, Hershberger RE, Day SM, Klinedinst NJ, Landstrom AP, Parikh VN, et al. Genetic testing for inherited cardiovascular diseases: a scientific statement from the American Heart Association. Circ Genom Precis Med. 2020:HCG0000000000000067. 10.1161/HCG.0000000000000067. This document provides expert consensus recommendations for genetic testing in individuals with inherited cardiovascular disease, including cardiomyopathy, arrhythmia, vascular disorders, and familial hypercholesterolemia. [DOI] [PubMed]
- 43.Miller IM, Lewis KL, Lawal TA, Ng D, Johnston JJ, Biesecker BB, et al. Health behaviors among unaffected participants following receipt of variants of uncertain significance in cardiomyopathy-associated genes. Genet Med. 2019;21(3):748–752. doi: 10.1038/s41436-018-0083-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scherr CL, Aufox S, Ross AA, Ramesh S, Wicklund CA, Smith M. What people want to know about their genes: a critical review of the literature on large- scale genome sequencing studies. Healthcare (Basel). 2018;6(3). 10.3390/healthcare6030096. [DOI] [PMC free article] [PubMed]
- 45.Tsai GJ, Ranola JMO, Smith C, Garrett LT, Bergquist T, Casadei S, et al. Outcomes of 92 patient-driven family studies for reclassification of variants of uncertain significance. Genet Med. 2019;21(6):1435–1442. doi: 10.1038/s41436-018-0335-7. [DOI] [PubMed] [Google Scholar]
- 46.Herman DS, Lam L, Taylor MR, Wang L, Teekakirikul P, Christodoulou D, et al. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366(7):619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haggerty CM, Damrauer SM, Levin MG, Birtwell D, Carey DJ, Golden AM, Hartzel DN, Hu Y, Judy R, Kelly MA, Kember RL, Lester Kirchner H, Leader JB, Liang L, McDermott-Roe C, Babu A, Morley M, Nealy Z, Person TN, Pulenthiran A, Small A, Smelser DT, Stahl RC, Sturm AC, Williams H, Baras A, Margulies KB, Cappola TP, Dewey FE, Verma A, Zhang X, Correa A, Hall ME, Wilson JG, Ritchie MD, Rader DJ, Murray MF, Fornwalt BK, Arany Z, On behalf of the DiscovEHR and Penn Medicine Biobank Studies Genomics-first evaluation of heart disease associated with titin-truncating variants. Circulation. 2019;140(1):42–54. doi: 10.1161/CIRCULATIONAHA.119.039573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Golbus JR, Puckelwartz MJ, Fahrenbach JP, Dellefave-Castillo LM, Wolfgeher D, McNally EM. Population-based variation in cardiomyopathy genes. Circ Cardiovasc Genet. 2012;5(4):391–399. doi: 10.1161/CIRCGENETICS.112.962928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verdonschot JAJ, Hazebroek MR, Wang P, Sanders-van Wijk S, Merken JJ, Adriaansen YA, van den Wijngaard A, Krapels IPC, Brunner-la Rocca HP, Brunner HG, Heymans SRB. Clinical phenotype and genotype associations with improvement in left ventricular function in dilated cardiomyopathy. Circ Heart Fail. 2018;11(11):e005220. doi: 10.1161/CIRCHEARTFAILURE.118.005220. [DOI] [PubMed] [Google Scholar]
- 50.Tobita T, Nomura S, Fujita T, Morita H, Asano Y, Onoue K, Ito M, Imai Y, Suzuki A, Ko T, Satoh M, Fujita K, Naito AT, Furutani Y, Toko H, Harada M, Amiya E, Hatano M, Takimoto E, Shiga T, Nakanishi T, Sakata Y, Ono M, Saito Y, Takashima S, Hagiwara N, Aburatani H, Komuro I. Genetic basis of cardiomyopathy and the genotypes involved in prognosis and left ventricular reverse remodeling. Sci Rep. 2018;8(1):1998. doi: 10.1038/s41598-018-20114-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jansweijer JA, Nieuwhof K, Russo F, Hoorntje ET, Jongbloed JD, Lekanne Deprez RH, et al. Truncating titin mutations are associated with a mild and treatable form of dilated cardiomyopathy. Eur J Heart Fail. 2017;19(4):512–521. doi: 10.1002/ejhf.673. [DOI] [PubMed] [Google Scholar]
- 52.Luk K, Bakhsh A, Giannetti N, Elstein E, Lathrop M, Thanassoulis G, Engert JC. Recovery in patients with dilated cardiomyopathy with loss-of-function mutations in the titin gene. JAMA Cardiol. 2017;2(6):700–702. doi: 10.1001/jamacardio.2017.0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brauch KM, Karst ML, Herron KJ, de Andrade M, Pellikka PA, Rodeheffer RJ, Michels VV, Olson TM. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J Am Coll Cardiol. 2009;54(10):930–941. doi: 10.1016/j.jacc.2009.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo W, Schafer S, Greaser ML, Radke MH, Liss M, Govindarajan T, Maatz H, Schulz H, Li S, Parrish AM, Dauksaite V, Vakeel P, Klaassen S, Gerull B, Thierfelder L, Regitz-Zagrosek V, Hacker TA, Saupe KW, Dec GW, Ellinor PT, MacRae CA, Spallek B, Fischer R, Perrot A, Özcelik C, Saar K, Hubner N, Gotthardt M. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18(5):766–773. doi: 10.1038/nm.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Refaat MM, Lubitz SA, Makino S, Islam Z, Frangiskakis JM, Mehdi H, Gutmann R, Zhang ML, Bloom HL, MacRae CA, Dudley SC, Shalaby AA, Weiss R, McNamara DM, London B, Ellinor PT. Genetic variation in the alternative splicing regulator RBM20 is associated with dilated cardiomyopathy. Heart Rhythm. 2012;9(3):390–396. doi: 10.1016/j.hrthm.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maatz H, Jens M, Liss M, Schafer S, Heinig M, Kirchner M, Adami E, Rintisch C, Dauksaite V, Radke MH, Selbach M, Barton PJR, Cook SA, Rajewsky N, Gotthardt M, Landthaler M, Hubner N. RNA-binding protein RBM20 represses splicing to orchestrate cardiac pre-mRNA processing. J Clin Invest. 2014;124(8):3419–3430. doi: 10.1172/JCI74523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parikh VN, Caleshu C, Reuter C, Lazzeroni LC, Ingles J, Garcia J, et al. Regional variation in RBM20 causes a highly penetrant arrhythmogenic cardiomyopathy. Circ Heart Fail. 2019;12(3):e005371. doi: 10.1161/CIRCHEARTFAILURE.118.005371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hey TM, Rasmussen TB, Madsen T, Aagaard MM, Harbo M, Molgaard H, et al. Pathogenic RBM20-variants are associated with a severe disease expression in male patients with dilated cardiomyopathy. Circ Heart Fail. 2019;12(3):e005700. doi: 10.1161/CIRCHEARTFAILURE.118.005700. [DOI] [PubMed] [Google Scholar]
- 59.Rampazzo A, Nava A, Malacrida S, Beffagna G, Bauce B, Rossi V, Zimbello R, Simionati B, Basso C, Thiene G, Towbin JA, Danieli GA. Mutation in human desmoplakin domain binding to plakoglobin causes a dominant form of arrhythmogenic right ventricular cardiomyopathy. Am J Hum Genet. 2002;71(5):1200–1206. doi: 10.1086/344208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Norman M, Simpson M, Mogensen J, Shaw A, Hughes S, Syrris P, et al. Novel mutation in desmoplakin causes arrhythmogenic left ventricular cardiomyopathy. Circulation. 2005;112(5):636–642. doi: 10.1161/CIRCULATIONAHA.104.532234. [DOI] [PubMed] [Google Scholar]
- 61.Sen-Chowdhry S, Syrris P, Ward D, Asimaki A, Sevdalis E, McKenna WJ. Clinical and genetic characterization of families with arrhythmogenic right ventricular dysplasia/cardiomyopathy provides novel insights into patterns of disease expression. Circulation. 2007;115(13):1710–1720. doi: 10.1161/CIRCULATIONAHA.106.660241. [DOI] [PubMed] [Google Scholar]
- 62.Bhonsale A, Groeneweg JA, James CA, Dooijes D, Tichnell C, Jongbloed JD, et al. Impact of genotype on clinical course in arrhythmogenic right ventricular dysplasia/cardiomyopathy-associated mutation carriers. Eur Heart J. 2015;36(14):847–855. doi: 10.1093/eurheartj/ehu509. [DOI] [PubMed] [Google Scholar]
- 63.Lopez-Ayala JM, Gomez-Milanes I, Sanchez Munoz JJ, Ruiz-Espejo F, Ortiz M, Gonzalez-Carrillo J, et al. Desmoplakin truncations and arrhythmogenic left ventricular cardiomyopathy: characterizing a phenotype. Europace. 2014;16(12):1838–1846. doi: 10.1093/europace/euu128. [DOI] [PubMed] [Google Scholar]
- 64.Reichl K, Kreykes SE, Martin CM, Shenoy C. Desmoplakin variant-associated arrhythmogenic cardiomyopathy presenting as acute myocarditis. Circ Genom Precis Med. 2018;11(12):e002373. doi: 10.1161/CIRCGEN.118.002373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh SM, Casey SA, Berg AA, Abdelhadi RH, Katsiyiannis WT, Bennett MK, Mackey-Bojack S, Duncanson ER, Sengupta JD. Autosomal-dominant biventricular arrhythmogenic cardiomyopathy in a large family with a novel in-frame DSP nonsense mutation. Am J Med Genet A. 2018;176(7):1622–1626. doi: 10.1002/ajmg.a.38719. [DOI] [PubMed] [Google Scholar]
- 66.Augusto JB, Eiros R, Nakou E, Moura-Ferreira S, Treibel TA, Captur G, Akhtar MM, Protonotarios A, Gossios TD, Savvatis K, Syrris P, Mohiddin S, Moon JC, Elliott PM, Lopes LR. Dilated cardiomyopathy and arrhythmogenic left ventricular cardiomyopathy: a comprehensive genotype-imaging phenotype study. Eur Heart J Cardiovasc Imaging. 2020;21(3):326–336. doi: 10.1093/ehjci/jez188. [DOI] [PubMed] [Google Scholar]
- 67.Ortiz-Genga MF, Cuenca S, Dal Ferro M, Zorio E, Salgado-Aranda R, Climent V, et al. Truncating FLNC mutations are associated with high-risk dilated and arrhythmogenic cardiomyopathies. J Am Coll Cardiol. 2016;68(22):2440–2451. doi: 10.1016/j.jacc.2016.09.927. [DOI] [PubMed] [Google Scholar]
- 68.Begay RL, Tharp CA, Martin A, Graw SL, Sinagra G, Miani D, Sweet ME, Slavov DB, Stafford N, Zeller MJ, Alnefaie R, Rowland TJ, Brun F, Jones KL, Gowan K, Mestroni L, Garrity DM, Taylor MRG. FLNC gene splice mutations cause dilated cardiomyopathy. JACC Basic Transl Sci. 2016;1(5):344–359. doi: 10.1016/j.jacbts.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nozari A, Aghaei-Moghadam E, Zeinaloo A, Mollazadeh R, Majnoon MT, Alavi A, Ghasemi Firouzabadi S, Mohammadzadeh A, Banihashemi S, Nikzaban M, Najmabadi H, Behjati F. A novel splicing variant in FLNC gene responsible for a highly penetrant familial dilated cardiomyopathy in an extended Iranian family. Gene. 2018;659:160–167. doi: 10.1016/j.gene.2018.03.044. [DOI] [PubMed] [Google Scholar]
- 70.Deo RC, Musso G, Tasan M, Tang P, Poon A, Yuan C, Felix JF, Vasan RS, Beroukhim R, de Marco T, Kwok PY, MacRae CA, Roth FP. Prioritizing causal disease genes using unbiased genomic features. Genome Biol. 2014;15(12):534. doi: 10.1186/s13059-014-0534-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Golbus JR, Puckelwartz MJ, Dellefave-Castillo L, Fahrenbach JP, Nelakuditi V, Pesce LL, Pytel P, McNally EM. Targeted analysis of whole genome sequence data to diagnose genetic cardiomyopathy. Circ Cardiovasc Genet. 2014;7(6):751–759. doi: 10.1161/CIRCGENETICS.113.000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Begay RL, Graw SL, Sinagra G, Asimaki A, Rowland TJ, Slavov DB, Gowan K, Jones KL, Brun F, Merlo M, Miani D, Sweet M, Devaraj K, Wartchow EP, Gigli M, Puggia I, Salcedo EE, Garrity DM, Ambardekar AV, Buttrick P, Reece TB, Bristow MR, Saffitz JE, Mestroni L, Taylor MRG. Filamin C truncation mutations are associated with arrhythmogenic dilated cardiomyopathy and changes in the cell-cell adhesion structures. JACC Clin Electrophysiol. 2018;4(4):504–514. doi: 10.1016/j.jacep.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ader F, De Groote P, Reant P, Rooryck-Thambo C, Dupin-Deguine D, Rambaud C, et al. FLNC pathogenic variants in patients with cardiomyopathies: prevalence and genotype-phenotype correlations. Clin Genet. 2019;96(4):317–329. doi: 10.1111/cge.13594. [DOI] [PubMed] [Google Scholar]
- 74.Verdonschot JAJ, Vanhoutte EK, Claes GRF. Helderman-van den Enden a, Hoeijmakers JGJ, Hellebrekers D, et al. A mutation update for the FLNC gene in myopathies and cardiomyopathies. Hum Mutat. 2020;41(6):1091–1111. doi: 10.1002/humu.24004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Landry LG, Rehm HL. Association of racial/ethnic categories with the ability of genetic tests to detect a cause of cardiomyopathy. JAMA Cardiol. 2018;3(4):341–345. doi: 10.1001/jamacardio.2017.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pottinger TD, Puckelwartz MJ, Pesce LL, Robinson A, Kearns S, Pacheco JA, Rasmussen-Torvik LJ, Smith ME, Chisholm R, McNally EM. Pathogenic and uncertain genetic variants have clinical cardiac correlates in diverse biobank participants. J Am Heart Assoc. 2020;9(3):e013808. doi: 10.1161/JAHA.119.013808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Villard E, Perret C, Gary F, Proust C, Dilanian G, Hengstenberg C, Ruppert V, Arbustini E, Wichter T, Germain M, Dubourg O, Tavazzi L, Aumont MC, DeGroote P, Fauchier L, Trochu JN, Gibelin P, Aupetit JF, Stark K, Erdmann J, Hetzer R, Roberts AM, Barton PJ, Regitz-Zagrosek V, Cardiogenics Consortium. Aslam U, Duboscq-Bidot L, Meyborg M, Maisch B, Madeira H, Waldenström A, Galve E, Cleland JG, Dorent R, Roizes G, Zeller T, Blankenberg S, Goodall AH, Cook S, Tregouet DA, Tiret L, Isnard R, Komajda M, Charron P, Cambien F. A genome-wide association study identifies two loci associated with heart failure due to dilated cardiomyopathy. Eur Heart J. 2011;32(9):1065–1076. doi: 10.1093/eurheartj/ehr105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meder B, Ruhle F, Weis T, Homuth G, Keller A, Franke J, et al. A genome-wide association study identifies 6p21 as novel risk locus for dilated cardiomyopathy. Eur Heart J. 2014;35(16):1069–1077. doi: 10.1093/eurheartj/eht251. [DOI] [PubMed] [Google Scholar]
- 79.Xu H, Dorn GW 2nd, Shetty A, Parihar A, Dave T, Robinson SW, et al. A genome-wide association study of idiopathic dilated cardiomyopathy in African Americans. J Pers Med. 2018;8(1). 10.3390/jpm8010011. [DOI] [PMC free article] [PubMed]
- 80.Aragam KG, Natarajan P. Polygenic scores to assess atherosclerotic cardiovascular disease risk: clinical perspectives and basic implications. Circ Res. 2020;126(9):1159–1177. doi: 10.1161/CIRCRESAHA.120.315928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Turner H, Jackson L. Evidence for penetrance in patients without a family history of disease: a systematic review. Eur J Hum Genet. 2020;28(5):539–550. doi: 10.1038/s41431-019-0556-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guo MH, Plummer L, Chan YM, Hirschhorn JN, Lippincott MF. Burden testing of rare variants identified through exome sequencing via publicly available control data. Am J Hum Genet. 2018;103(4):522–534. doi: 10.1016/j.ajhg.2018.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Minikel EV, Vallabh SM, Lek M, Estrada K, Samocha KE, Sathirapongsasuti JF, et al. Quantifying prion disease penetrance using large population control cohorts. Sci Transl Med. 2016;8(322):322ra9. doi: 10.1126/scitranslmed.aad5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ramchand J, Wallis M, Macciocca I, Lynch E, Farouque O, Martyn M, Phelan D, Chong B, Lockwood S, Weintraub R, Thompson T, Trainer A, Zentner D, Vohra J, Chetrit M, Hare DL, James P. Prospective evaluation of the utility of whole exome sequencing in dilated cardiomyopathy. J Am Heart Assoc. 2020;9(2):e013346. doi: 10.1161/JAHA.119.013346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mak TSH, Lee YK, Tang CS, Hai JSH, Ran X, Sham PC, et al. Coverage and diagnostic yield of whole exome sequencing for the evaluation of cases with dilated and hypertrophic cardiomyopathy. Sci Rep. 2018;8(1):10846. doi: 10.1038/s41598-018-29263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Minoche AE, Horvat C, Johnson R, Gayevskiy V, Morton SU, Drew AP, Woo K, Statham AL, Lundie B, Bagnall RD, Ingles J, Semsarian C, Seidman JG, Seidman CE, Dinger ME, Cowley MJ, Fatkin D. Genome sequencing as a first-line genetic test in familial dilated cardiomyopathy. Genet Med. 2019;21(3):650–662. doi: 10.1038/s41436-018-0084-7. [DOI] [PMC free article] [PubMed] [Google Scholar]