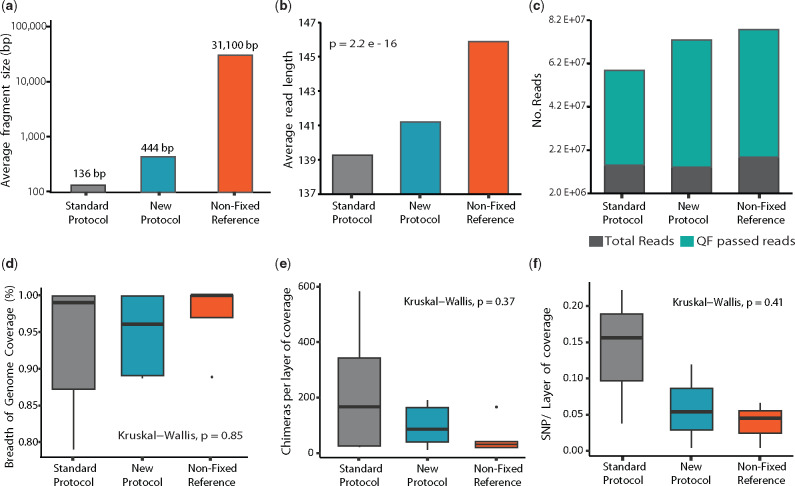
Figure 6:
Combined protocol–bacterial DNA. Outputs of Bioanalyzer and WGS for bacterial FFPE DNA exposed to the combined treatment (blue, labelled as new protocol, Σn = 6). This was compared with that obtained from six pooled paired-samples decrosslinked with the reference protocol and unrepaired (grey, labelled reference protocol, Σn = 6) and that from DNA obtained from NF samples with the same bacterial and DNA content (orange, labelled NF, Σn = 3). Improvement in DNA readability, sequence quality and integrity was measured by Integrity (fragment length): (a) fragment analyser (b) WGS. Readability: (c) Quantity of reads and filter pass reads per coverage. (d) Percentage of breath of genome coverage. Sequence quality: (e) number of chimeric reads per layer of coverage. (f) Number of SNPs per layer of coverage.