FIGURE 2.
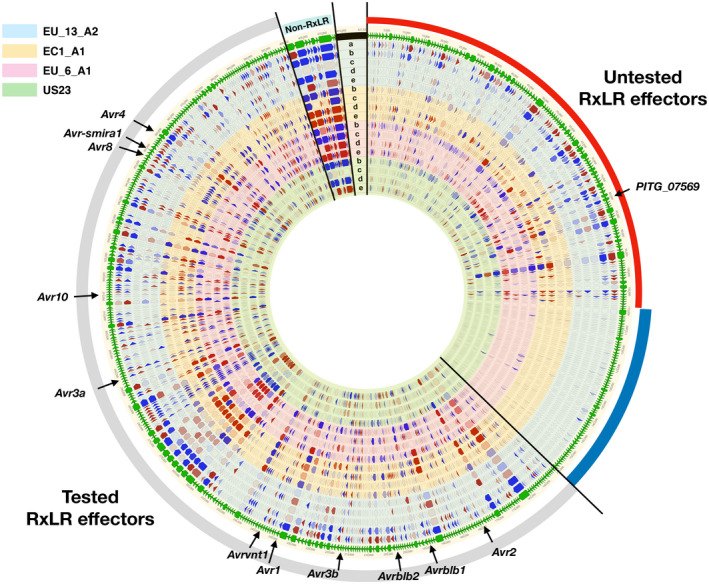
cDNA pathogen‐enrichment sequencing (PenSeq) of RXLR effectors from EU_13_A2, EC1_A1, EU_6_A1, and US23. The cDNA PenSeq data for the RXLR effectors from four Phytophthora infestans at different stages were mapped to an artificial contig (RXLRome) of 499 RXLR effectors and nine non‐RXLR genes, demarcated by bright green arrows on the outer edge of the diagram. Black lines separate the previously tested RXLR effectors (grey bar), new effector candidates with differential expression (red bar), unexpressed effectors (blue bar) and non‐RXLR controls (cyan). The concentric circles in blue, yellow, pink, and green represent data from P. infestans EU_13_A2, EC1_A1, EU_6_A1, and US23, respectively. The arrows on them indicate differential expression (red, up‐regulation; blue, down‐regulation; no fill, no difference), where the more intense the colour, the bigger the difference. The data are plotted as follows: a, mycelium vs. zoospores (for EU_13_A2 only); b, zoospores vs. 12 hr postinoculation (hpi); c, 12 hpi vs. 1 day postinoculation (dpi); d, 1 dpi vs. 2 dpi; e, 2 dpi vs. 3 dpi. Eleven known Avr genes, namely Avr4, AvrSmira1, Avr8, Avr10, Avr3a, Avrvnt1, Avr1, Avr3b, Avrblb2, Avrblb1, and Avr2, are indicated by black arrows. PITG_07569 is indicated by a black arrow
