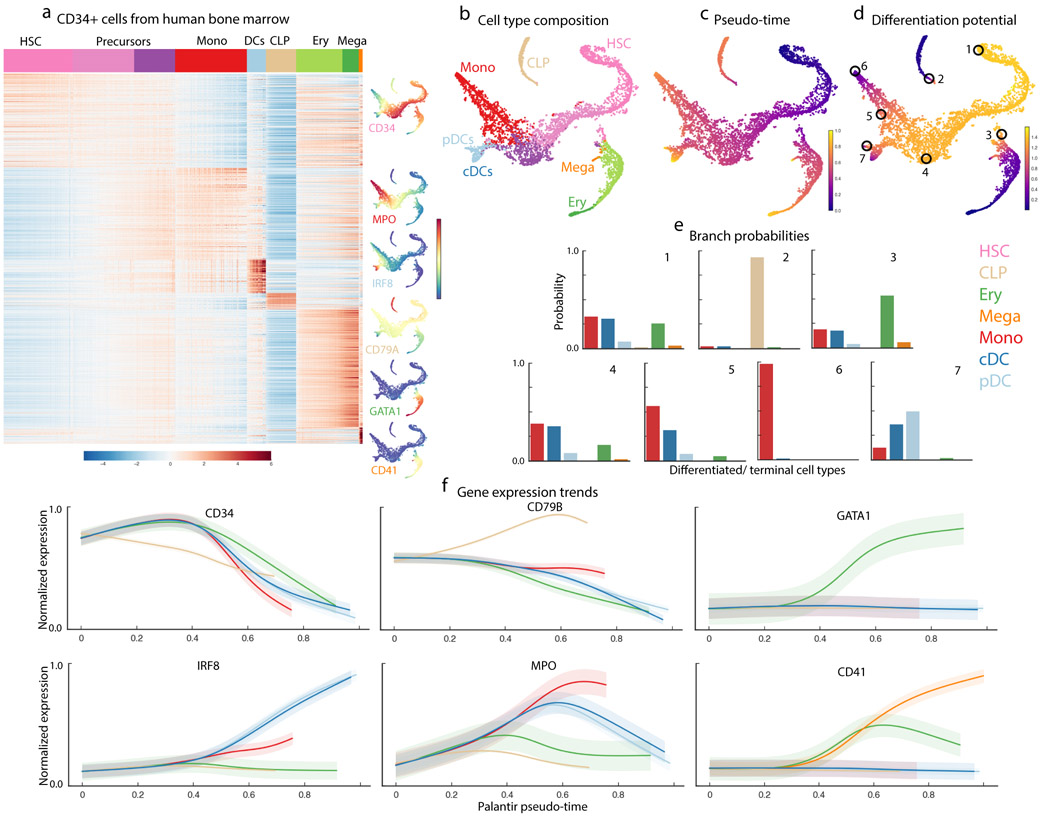
Figure 2.
Differentiation landscape of early human hematopoiesis. Data shown for CD34+ human bone marrow cells, replicate 1. (a) MAGIC imputed expression of genes (rows) differentially expressed between PhenoGraph13 clusters (based on MAST51). Cells (columns) are ordered by cluster; top row represents annotated cluster labels, with color coding scheme used in all figures. tSNE maps show cells colored by imputed expression of characteristic cell lineage markers. (b-d) tSNE maps of full scRNA-seq dataset generated using one HSC as a start cell. 5780 cells are shown on the tSNE maps.(b) Cells colored by cluster labels in (a), annotated by correlation with bulk sorted populations. (c) Cells colored by Palantir pseudo-time. (d) Cells colored by Palantir differentiation potential. (e) Branch probabilities of example cells circled in (d), highlighting early cells (1), lymphoid and erythroid lineages (2,3) and monocyte and DC lineages (4-7). Bars are colored by cell type as in (a). (f) Gene expression trends for characteristic lineage genes, plotted as in Supplementary Fig. 3.