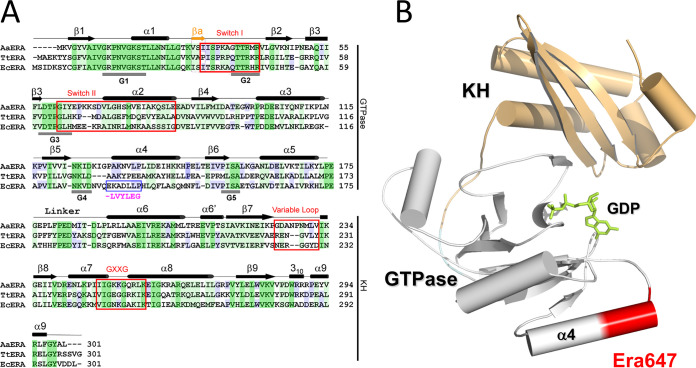
FIG 1.
The structure of the Era protein and location of the Era647 mutation. (A) A comparison of amino acid sequences among Era homologs from Aquifex aeolicus, Thermus thermophilus, and E. coli (PDB entries 3IEV, 1WF3, and 1EGA, respectively) is shown along with structural features. Identical residues shared among all three homologs are shaded dark green, similar residues shared among all three homologs are shaded light green, and identical residues shared with two homologs are shaded blue. Residues 131 to 137 changed in Era647 are highlighted with a blue box and magenta letters below; the deleted glutamate is shown as a dash. (B) The location of residues 131 to 137 near α helix 4 of the GTPase domain is indicated on the atomic structure of E. coli Era in complex with GDP (PDB entry 3IEU). The ribbon diagram represents the protein (helices as cylinders, strands as arrows, and loops as tubes), and the stick model in green represents the nucleotide. The GTPase and KH domains are colored in gray and light orange, respectively, and residues 131 to 137 are highlighted in red.