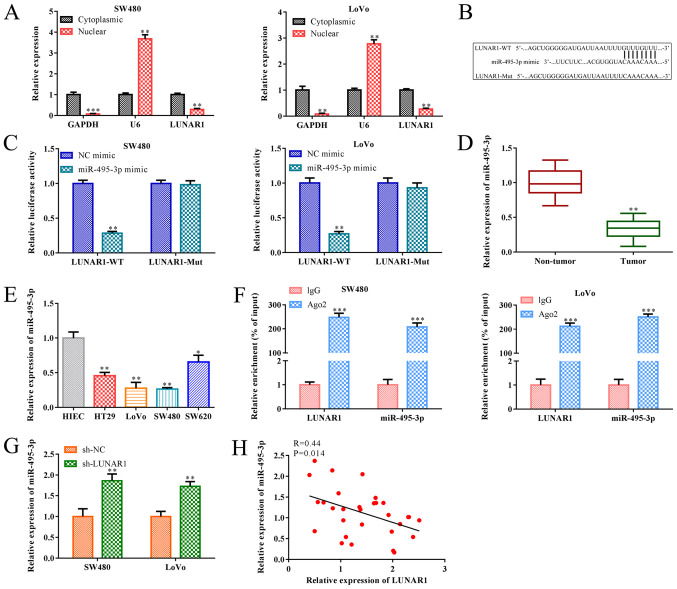
Figure 4.
LUNAR1 functions as a sponge of miR-495-3p. Cytoplasm and nuclear grades were separated from SW480 and LoVo cells. (A) RT-qPCR was performed to assess the levels of U6 and LUNAR1 in the cytoplasm and nuclei. **P<0.01, ***P<0.001 vs. cytoplasmic LUNAR1. (B) Potential binding site between miR-495-3p and LUNAR1 was predicted by TargetScan software. miR-495-3p mimics were transfected into SW480 and LoVo cells for 24 h; NC mimics served as a control. (C) Dual luciferase reporter gene assay was performed to confirm the direct binding relationship between miR-495-3p and LUNAR1. **P<0.01 vs. the NC mimic group. (D) RT-qPCR was carried to detect the RNA level of miR-495-3p in CRC and non-tumor tissues. **P<0.01 vs. the non-tumor tissues. (E) RT-qPCR was carried to detect the RNA level of miR-495-3p in CRC cell lines; HIEC cells served as the control. *P<0.05, **P<0.01 vs. the HIEC cells. (F) Interaction between LUNAR1 and miR-495-3p was determined by RIP assay. ***P<0.001 vs. the IgG group. SW480 and LoVo cells were transfected with sh-LUNAR1 for LUNAR1 knockdown for 24 h, and sh-NC served as a negative control. (G) RT-qPCR was performed to assess miR-495-3p RNA levels. **P<0.01 vs. the sh-NC group. (H) Pearson's correlation analysis was conducted to analyze the correlation between LUNAR1 and miR-495-3p in SW480 and LoVo cells. Comparisons were performed using paired t-test or one-way ANOVA. Error bars represent SD. Data represent three independent experiments. LUNAR1, lncRNA LUNAR1; CRC, colorectal cancer.