Figure 3.
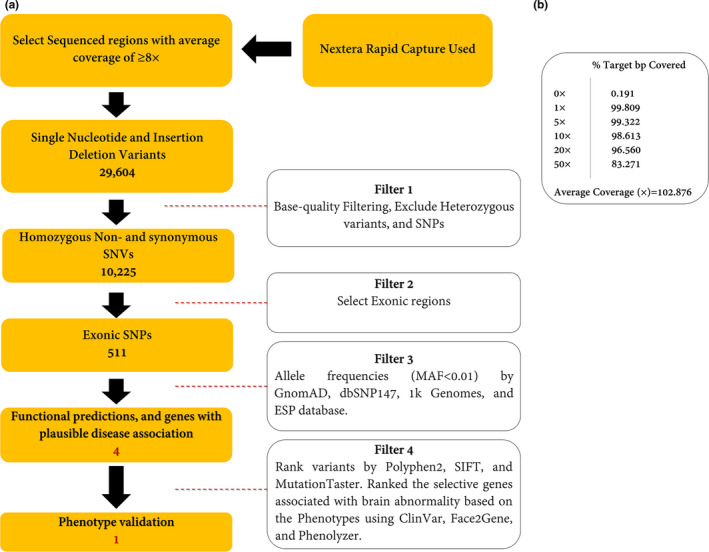
(a) Variant filtering scheme used to prioritize the variants of exome data. Approximately 37 Mb (214,405 exons) of the Consensus Coding Sequences (CCS) were enriched from fragmented genomic DNA by >340,000 probes designed against the human genome (Nextera Rapid Capture Exome, Illumina) and the generated library sequenced on an Illumina platform to an average coverage depth ~100×. The regions with an average coverage of ≥8× were selected. Other procedures are discussed in the main text in detail. (b) Analysis of statistics WES. The average coverage was about 102.876×
