Figure 4.
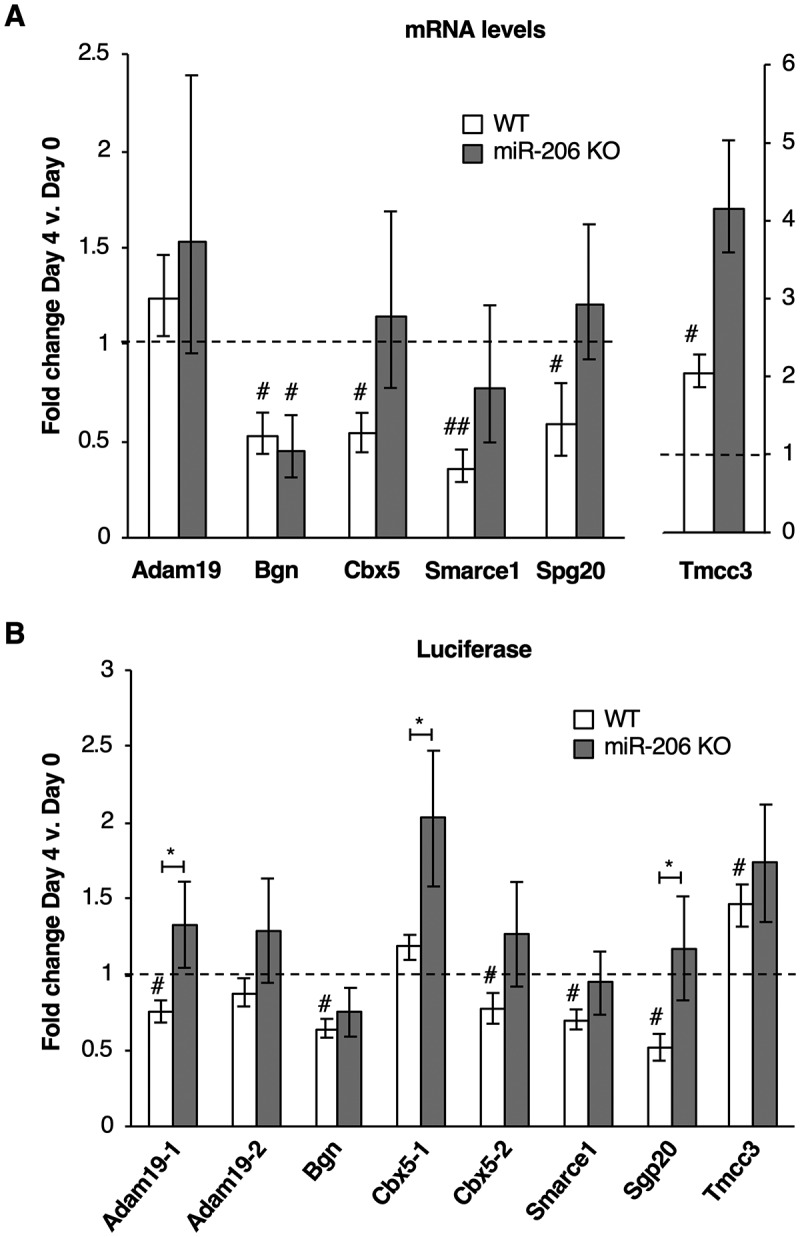
Putative mRNA targets of miR-206 identified by xOP-seq are differentially regulated in miR-206 KO cells compared to WT cells. (A) Endogenous levels of Bgn, Cbx5, Smarce1, and Spg20 mRNAs decrease in WT cells upon differentiation and show different levels of regulation in miR-206 KO cells after four days in differentiation media. Normalized average fold change in mRNA expression on Day 4 relative to Day 0 is plotted. The error bars denote standard deviations (n = 3). The dashed line at 1.0 represents no change in mRNA levels. # indicates a p-value < 0.05 and ## indicates a p-value <0.01 as compared to 1.0 using a one sample two-tailed t-test, except for Spg20 in WT, which has a p-value < 0.068. (B) Regions from the 3ʹUTRs of five putative miR-206 target mRNAs (Adam19-1, Bgn, Cbx5-2, Smarce1 and Spg20) downregulate luciferase expression in WT cells but not miR-206 KO cells after treatment with differentiation media. Normalized average fold change in luciferase expression after differentiation is plotted; the error bars denote standard deviations (n = 3). The dashed line at 1.0 represents no change in luciferase expression. # indicates a p-value < 0.05 as compared to 1.0 using a one sample two-tailed t-test, except for Cbx5-2 in WT, which has a p-value < 0.062. * indicates a p-value < 0.05, as determined from an unpaired two-tailed t-test comparing the data from the WT and KO cells.
