Figure 1.
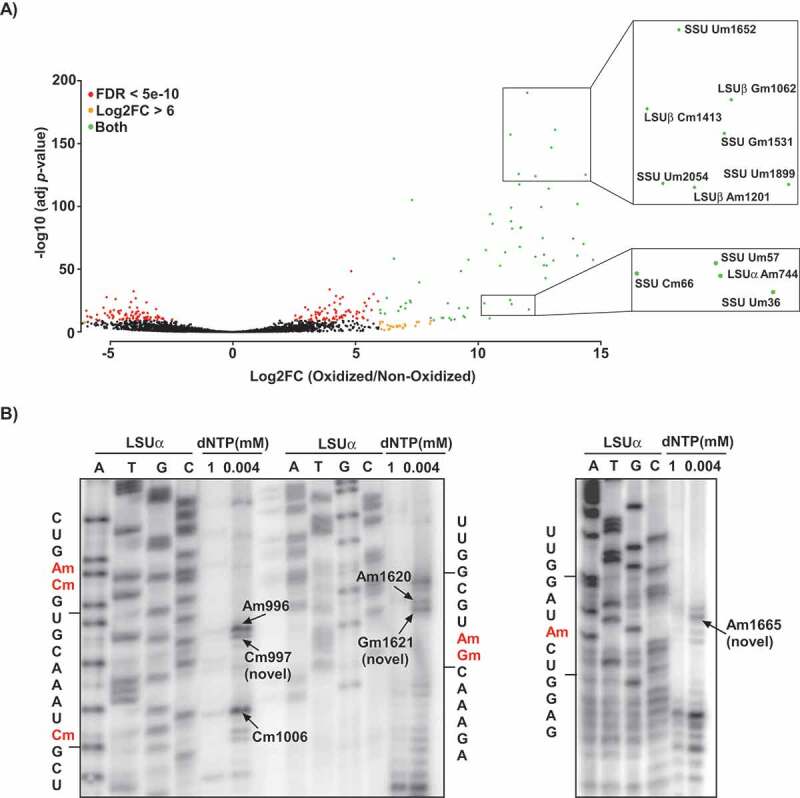
Detection of Nm positions in T. brucei rRNA using RibOxi-seq. A) Volcano plot illustrating the detection of Nm positions in T. brucei rRNA. Total RNA from T. brucei PCF was treated (oxidized) with NaIO4, adapters or left un-treated (non-oxidized) and ligated and sequenced as described in ‘Material and Methods’. The reads were mapped to the rRNA, and the positions having a cut-off value of log2FC >6 and adjusted P-value of <10−11 are presented. The X-axis represents the log2FC (oxidized/non-oxidized) and the Y-axis represents the -log10 (P-value) across each site detected in the rRNA. Positions meeting the criteria of log2FC>6 are indicated as orange dots, those meeting FDR < 5e-10 are indicated by red dots, and positions qualifying both log2FC and FDR are shown in green. The zoomed-in views indicate examples of the detected methylation sites. B) Validation of novel Nm sites detected by RibOxi-seq. Total RNA (10 μg) was subjected to primer extension using an oligonucleotide complementary to an indicated rRNA region at low and high-dNTP concentrations (0.004 and 1 mM, respectively). Extension products were separated on 10% polyacrylamide–7 M urea gels along with a dideoxynucleotide sequence ladder of the rRNA produced using the same primer. Partial RNA sequences are depicted, and the methylated nucleotide is highlighted in red. Locations of the Nms on rRNA are indicated.
