Figure 5.
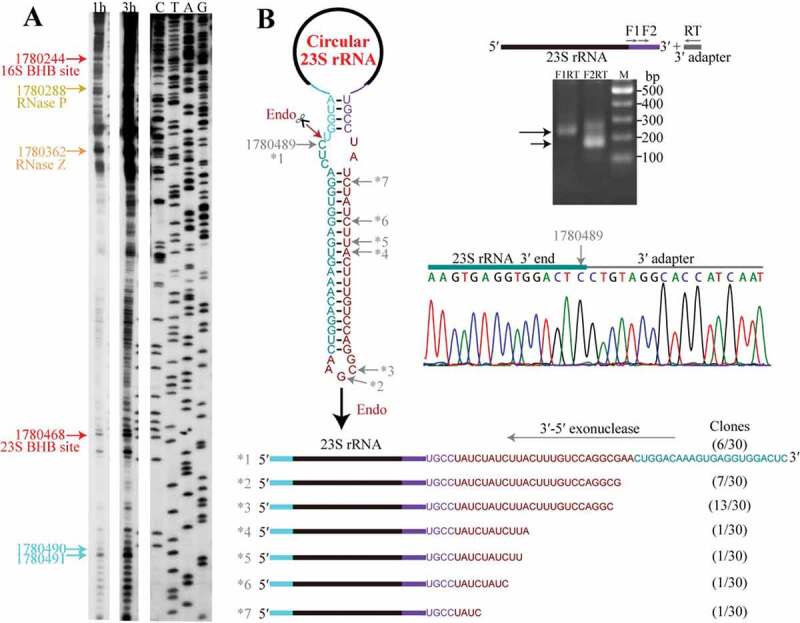
Linearization of the circular pre-23S rRNA intermediate followed by 3′-5′ exoribonucleolytic trimming. (a) Primer extension detected the major processing sites upstream of 23S rRNA. Cyan arrows indicate the two mature 5′ ends of 23S rRNA, and the upstream major processing sites are indicated by the same coloured arrows shown in Fig. 1. 1 h and 3 h at gel top indicate autographing times. (b) 3′-RACE assayed the 3′-5′ exoribonucleolytic processing trace in pre-23S rRNA maturation. A diagram of the circular pre-23S rRNA shows the complete precursor sequence (left upper panel) with the featured RNA fragments coloured same as in Figure 3(a). An endoribonucleolytic processing event (dark red arrow) was predicted to linearize the circular pre-23S rRNA. 3′-RACE was performed as depicted in the right upper panel and the products were sequenced. The nested PCR products in 3′-RACE were analysed on 2% agarose gel (right upper panel) and the DNA sequencing results of nt 1780489 (right lower panel) are shown. The remaining six representative 3′ ends (*2 to *7) captured by 3′-RACE are shown as a diagram (lower panel), and the corresponding sites are indicated in the sequence at the upper panel. Numbers in parenthesis indicate the sequenced clones.
