Figure 8.
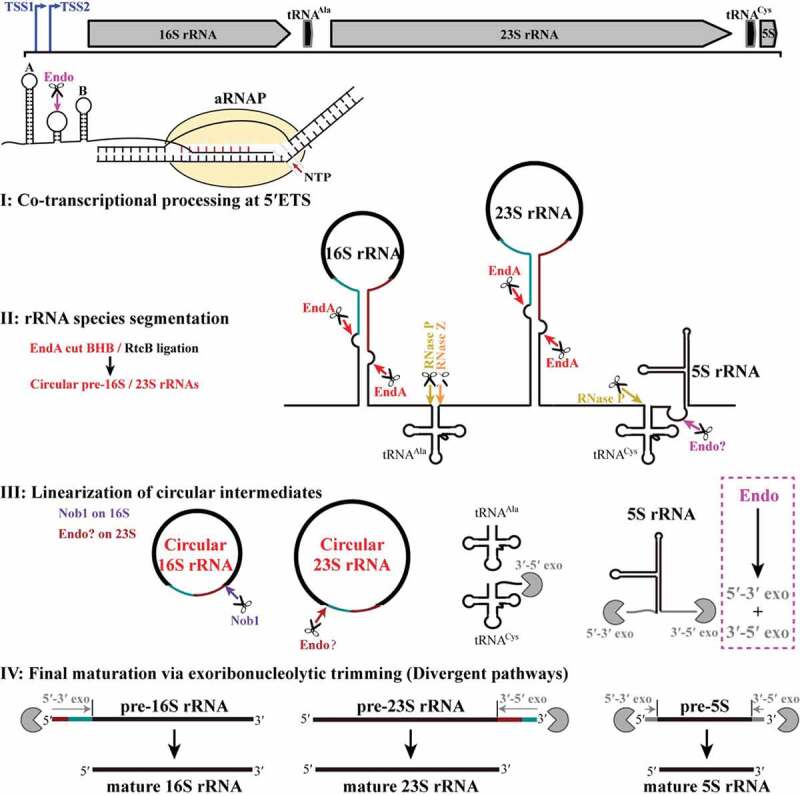
The pre-rRNA processing and maturation mode in M. psychrophilus. The rRNA operon (16S-tRNAAla-23S-tRNACys-5S) is transcribed as a polycistronic RNA from two transcription start sites (TSS1 and TSS2). A co-transcriptional endoribonucleolytic cleavage (Endo) at 5′ ETS that removes helix A initiates the processing (Step I). Following pre-rRNA transcription and folding, rRNAs and tRNAs are segmented (Step II) by the corresponding endoribonucleases. Splicing endonuclease EndA cleaves at BHB motifs buried in the processing stems to excise pre-16S and pre-23S rRNAs and RNA ligase RtcB catalyzes the coupled circularization. Cleavages of RNase P and RNase Z release tRNAAla, while cleavages of RNase P and an unknown endoribonuclease separate tRNACys and 5S rRNA. The circular pre-16S and pre-23S rRNA intermediates are linearized (Step III). Nob1 linearizes the circular 16S rRNA at the mature 3′ end and generates a long 5′ extension. In contrast, an unknown endoribonuclease linearizes the circular 23S rRNA at the mature 5′ end and generates a short 3′ extension. Finally, linearized pre-16S, 23S, and 5S rRNAs are trimmed through different exoribonucleolytic pathways (Step IV). The 5′ and 3′ extensions of pre-16S and pre-23S rRNAs are finally matured by 5′-3′ and 3′-5′ exoribonucleolytic trimming, respectively, while exoribonucleolytic trimming of both ends are involved in 5S rRNA maturation. The scissor and Pacman icons represent an endoribonuclease and exoribonuclease, respectively. Dark cyan and red lines at pre-16S and pre-23S rRNAs show the precursor regions in circular intermediates. Magenta dotted line frames a postulated pre-5S rRNA maturation mode by a combination of endoribonucleolysis and exoribonucleolysis.
