Figure 4.
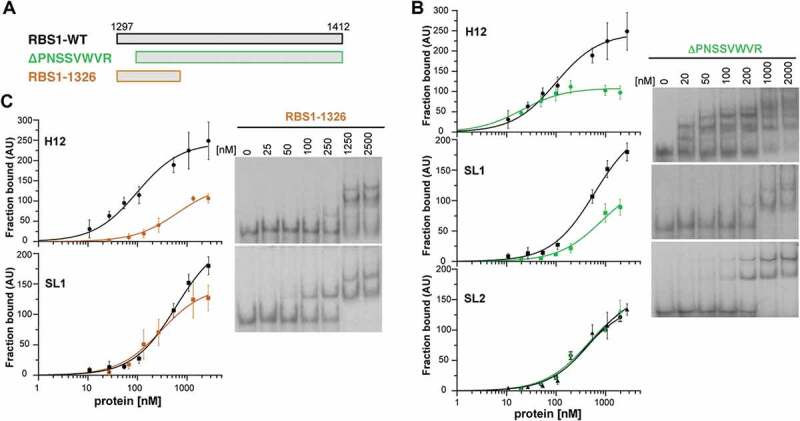
RBS1 constructs carrying deletions of the N-terminal or C-terminal residues exhibit different RNA-binding capacity. (A) Schematic representing different versions of His-RBS1 protein used in gel-shift analysis. (B) Graph representing the adjusted curves obtained from the quantification (mean ± SD) of three independent gel-shift assays using H12, SL1, or SL2 probes incubated with increasing amounts of RBS1-WT (black line) or RBS1ΔPNSSVWVR (green line) proteins. Representative examples of the gel-shift assays conducted with labelled H12, SL1, or SL2 with RBS1ΔPNSSVWVR probes (right panel). (C) Graph representing the adjusted curves obtained from the quantification (mean ± SD) of independent assays using H12 or SL1 probes incubated with increasing amounts of RBS1-WT (black line) or RBS11326 (brown line) proteins. Representative examples of the gel-shift assays conducted with labelled H12 or SL1 probes with RBS11326 (right panel).
