Figure 5.
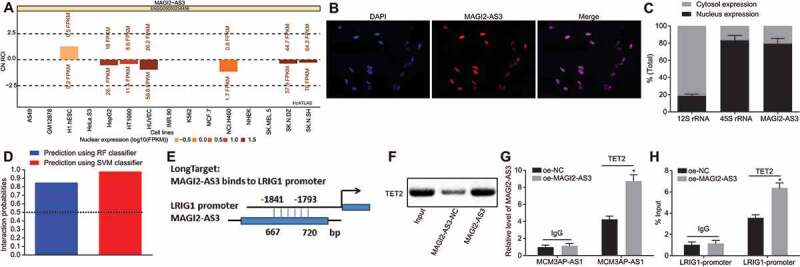
LncRNA MAGI2-AS3 promotes DNA demethylation of LRIG1 by recruiting TET2. A: lncATLAS identifies the cellular localization of MAGI2-AS3 in nucleus; B: The cellular localization of MAGI2-AS3 is verified using FISH; C: The cellular localization of MAGI2-AS3 in nucleus is confirmed using fractionation of nuclear and cytoplasmic RNA; D: RPIseq databank predicts DNA demethylase TET2 are relevant to MAGI2-AS3 using RF classifier and SVM classifier (RF > 0.5 & SVM > 0.5); E: potential binding sites for MAGI2-AS3 within LRIG1 promoter region predicted by LongTarget; F: RNA pull-down shows TET2 binds to MAGI2-AS3 using western blot analysis; G: binding of MAGI2-AS3 to TET2 in LSCs with MAGI2-AS3 overexpression in RIP assay followed by RT-qPCR; H: enrichment of TET2 in LRIG1 promoter region revealed by ChIP-qPCR; * p < 0.05 vs. the oe-NC group; data were expressed as mean ± standard error and analysed using unpaired t-test. The experiment was repeated three times independently.
