ABSTRACT
Dengue viruses (DENV) are the wildest transmitted arbovirus members of the family Flaviviridae, genus Flavivirus. Dengue viruses are composed of four serotypes, DENV1, 2, 3, and 4, and these viruses can cause dengue fever and dengue haemorrhagic fever or dengue shock syndrome, when infecting humans. RNA interference (RNAi) is a self-defence mechanism, which can be used to prevent invasions of RNA viruses to the host. Genetically engineering a host with an RNAi molecule that targets a single virus serotype may develop escape mutants, and can cause unusual dominance over other serotypes. Therefore, the simultaneous targeting of multiple serotypes is necessary to block DENV transmission. Here, we report the development of transgenic Aedes aegypti based on a bioinformatically designed multiple miRshRNA (microRNA-based shRNA) DNA sequence under the control of a blood-meal induced promoter, Carboxypeptidase A, to induce RNAi for DENV in Aedes aegypti, and demonstrate the expression of a synthetic multiple shRNA polycistronic cluster having RNA interference sequences to target DENV genomes. The transgenic mosquitoes have lower rates of infection, dissemination, and transmission for DENV2 and DENV4 compared to wild mosquitoes, with a significant reduction of dengue copy number and antigen levels in the midgut. These levels of DENV were low enough to make transgenic mosquitoes stop the DENV transmission from infected host to a susceptible host and refractory to DENV2 and DENV4 infection. Such multiple resistance in Ae. aegypti has not been documented previously. Laboratory fitness measurement of transgenic Ae. aegypti showed results comparable to other reported transgenic mosquitoes.
KEYWORDS: RNA interference, multiple miRshRNA, aedes aegypti, dengue disease, piggyBac
Introduction
Dengue (DEN) causes epidemics in more than 100 tropical and sub-tropical countries. It is a vector-borne disease caused by the mosquito, Aedes aegypti, the principal vector for epidemic dengue disease [1]. Dengue fever (DF) is caused by any of the four closely related serotypes of the dengue virus (DENV) (family Flaviviridae; genus Flavivirus). The infection causes febrile flu-like illness and occasionally develops into potentially lethal complications called dengue haemorrhagic fever (DHF) and dengue shock syndrome [2]. Over 2.5 billion people (over 30% of the world’s population) are now at risk of DENV infection. According to WHO reports, 50–100 million of the global population are infected annually by DENV [3]. Sri Lanka has been affected by epidemics of DF for over the past 2 decades. In recent years, DEN has become the number one vector-borne disease in Sri Lanka, and it experienced the worst ever DEN outbreaks in 2009 with 346 deaths from 35,000 cases, and in 2010 with 229 deaths from 32,292 cases [2]. Further, in 2013, 31,975 suspected DEN cases were reported showing that DEN is a major health issue in Sri Lanka. Dengue virus serotypes 2 (DENV2) and 3 (DENV3) were responsible for 86% of dengue infections until 2009. During this period dengue serotype 1 (DENV1) only accounted for 7% of infections. However, a study in 2010 reported that DENV1 accounted for more than 95% of DENV infections in Sri Lanka [4]. There is no specific treatment for DENV disease, and DENV prevention and control entirely depends on effective vector control measures, but vector control strategies that minimize human–mosquito contact have largely failed [5,6]. Control of the Ae. aegypti mosquito is extremely important because this mosquito transmits DF as well as other diseases such as yellow fever [7] and chikungunya [8]. Therefore, over the last few years, in addition to traditional mosquito control strategies, researchers have been developing more promising genetic-based strategies to produce transgenic mosquitoes that are capable of controlling mosquito-borne diseases [9–12]. These strategies include engineering mosquitoes with (i) impaired competency to transmit pathogens, based on RNA interference (RNAi) (ii) lethal genes, involving the release of insects carrying dominant lethal genes (RIDL) (iii) editing genes using the CRISPR/Cas9 tool to implement a gene-drive to replace the wild population. These strategies can steadily overcome the challenges imparted by mosquitoes to humans and other animals [13–15].
The strategies based on RNAi have been developed to achieve transgenic mosquitoes with impaired competency to transmit pathogens. This has been implemented to induce silencing of viral replication in mosquitoes using double-stranded RNA (dsRNA) or short hairpin RNA (shRNA) based mechanisms [9,16]. A key step towards developing this control strategy is to identify effector genes that inhibit dengue DENV replication when expressed in the mosquito vectors. Studies have shown that the extended dsRNA(567nt) derived from the prM protein-coding region of DENV2 could block the replication of this serotype in Aedes albopictus C6/36 cell lines [17] and subsequently, in the mosquito midgut epithelium, a site of an attack by the vulnerable stage virus [9]. Later in 2012, a study conducted by Korrapati and co-workers showed that RNAi carried out using the most suitable effecter DENV target sequences could knockout the majority of DENV in HEK293 cells by adenoviral-mediated delivery methods [18]. The main advantage of this shRNA-based method is the possibility of delivering many small engineered effector gene sequences to many targets of the viral genome. Another approach that extended to the shRNA-based method is microRNA (miRNA)-based shRNA (miRshRNA or second-generation shRNA), and this approach has been proven to be effective for HIV-1 in human embryonic kidney 293 FT adherent cells by strongly reducing the expression of the virus by creating multiple miRshRNA (multi-miRshRNA) targets to HIV-1 genome [19]. Therefore, designing such shRNAs as miRshRNA using the host miRNA sequence can ensure host-specific expression and increase the number of targets in the DENV genome. Hence, in this study, we designed a multi-miRshRNA molecule with a smaller size (~400 bp) compared to current approaches [16,20]. Multiple DENV serotype targeting from a particular miRshRNA in the design was achieved to reduce the size of the molecule and to overcome escape mutants as much as possible. The designed multi-miRshRNA that seems to be effective for DENV genome sequences of Sri Lankan origin were validated by constructing transgenic Ae. aegypti.
Results
Designing multiple microRNA-based shRNA
The DENV target sequences, microRNA-based shRNA (miRshRNA) in the transgene cassette, were selected from the two sequences having shown to be effective for DENV multiple serotypes (Korrapati et al, 2012, Table 1, Fig. 1A), and two conserved sequences identified in this study from the capsid region of DENV1 and the NS5 region of DENV3 of Sri Lankan origin (Figure S1). Ae. aegypti transcriptome-wide off-target analyses using the selected shRNA did not reveal any contiguous sequences of >17bp. Thus, in designing DENV target sequences, each of these miRshRNA sequences was placed within the Ae. aegypti microRNA-1175 (miRBase.org ref: MI0013470) backbone as shown in Fig. 1B and connected together to create the multiple miRsRNA molecule (multi-miRshRNA) (Fig. 1C) to have structures with four loops, having the lowest ΔG value in the mfold online RNA structure prediction application (Figure S2). This multi-miRshRNA was then connected to the downstream side of the Ae. aegypti Carboxypeptidase A (AeCPA) gene promoter to design the RNAi gene to achieve blood meal-inducible, midgut-specific expression of multi-miRshRNA. The designed RNAi gene (RNAiDENV), resistant to DENV in Ae. aegypti, comprises AeCPA, multi-miRshRNA, and the Simian virus 40 terminator sequence (svA) (Fig. 1D)
Table 1.
Chosen DENV target sequences in designing miRshRNAs.
| Name | Sequence (5ʹ-3ʹ) | Target Site | Element targeted | Target DENV Serotypes |
|---|---|---|---|---|
| miRK1.1 | AGACCAGAGATCCTGCTGTCT | 10,639–10,659 | 3ʹUAR | D1, D2, D3 & D4 |
| miRK2.6 | TTAGAGAGCAGATCTCTGATG | 79-99 | 5ʹUAR | D2, D3 and D4 |
| miR14.5 | AACTCTGGAGCAAATGCAAAG | 8,146–8,166 | NS5 | D1 |
| miR37.4 | AACCGTCTATCAATATGCTGA | 123- 143 | Capsid | D3 |
5ʹUAR- 5ʹ untranslated region, 3ʹUAR- 3ʹ untranslated region, NS5- nonstructural protein 5 and Capsid- capsid protein of dengue virus genome
Figure 1.
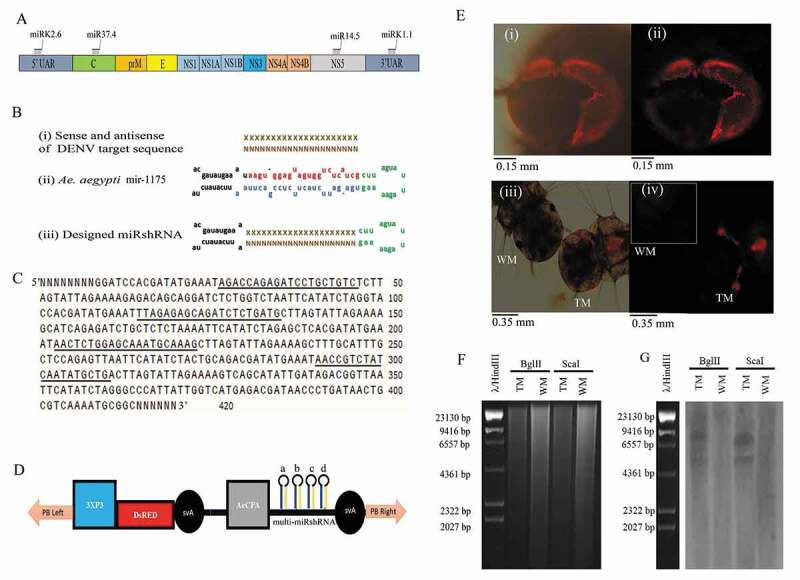
Trangenic construct design and transgene analysis in Ae. aegypti. (A) Schematic diagram of the DENV genome showing of structural proteins [capsid (C), membrane precursor (prM), and envelope (E)] and nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) and selected regions for dengue virus targets (Table 1). (B) Single miRshRNA design. (I) Sense of DENV target sequence shown in ‘XXXXX’ and antisense of DENV target sequence shown in ‘NNNNN’, (ii) Tail, mature sense, antisense and loop sequences of host-specific Ae. aegypti pre-microRNA, mir-1175 are shown in black, red, blue and green colour letters respectively, (iii) designed miRshRNA comprising tail of mir-1175, sense and antisense of DENV target sequences and mir-1175 loop. (C) Nucleotide sequence of designed multi-miRshRNA. Selected dengue virus target sequences are underlined in order (5ʹ-3ʹ) of miRK1.1, miRK2.6, miR14.5, and miR37.4 and random sequence shown in ‘NNNNN’. (D) Transgene construct containing 3xP3, DsRED, svA, RNAiDENV [AeCPA, miRshRNAs of DENV target sequences (a: miRK1.1, b: miRK2.6, c: miR14.5 and d: miR37.4) and svA sequence] and piggyBac Left(PB left) and piggyBac right (PB Right) sequences at 5ʹ and 3ʹ ends. (E) DsRED fluorescent protein expression in transgenic Ae. aegypti Sri Lankan strain. (i) Fluorescent image of the adult compound eye of Transgenic Mosquito (TM), (ii) Superimposed fluorescent image of the adult compound eye of TM. (iii) Fluorescent image of TM and wild mosquito (WM) larvae (iv) Superimposed fluorescent image of TM larvae and WM larvae (shown in white border). Fluorescence was visualized using Olympus BX53 with DsRED fluorescent protein filter. Magnification is 20X. (F) Agarose gel image of genomic DNA digestion of TM and WM, Genomic DNAs digested with restriction enzyme BglII or ScaI were separated in 0.8% agarose. (G) Southern blot analyses of DNA extracted from TM and WM. Restriction enzyme digested DNA blotted into the nylon membrane and hybridized it with Biotin-labelled DNA probes complementary to the sequence of the svA and piggyBac right sequence as described in the methodology. (AeCPA: midgut-specific Ae. aegypti carboxypeptidase A induction promoter, 3xP: Eye-specific promoter, DsRED: Red fluorescent protein, svA: Simian virus 40 terminator sequence, TM: transgenic mosquito, WM: wild mosquito, λ/HindIII: λ/HindIII maker, DENV: dengue virus, miRshRNA: microRNA based shRNA, multi-miRshRNA; multiple miRshRNA).
Engineering of transgenic mosquitoes
The cargo plasmid containing the piggyBac transposable element and eye-specific reporter gene [21] that drives the expression of DsRED protein (Red fluorescent protein) in ommatidium units as a transgenesis marker was used to introduce RNAiDENV into the Ae. aegypti via microinjection. The 231 lines of G1 mosquitoes that survived were screened for DsRED protein expression in the ommatidium units of their eyes (Fig. 1E), and six lines were shown to have the DsRED expression in their eyes. Out of these, the transgenic line TRSL1161 was selected for further studies, and was maintained as hemizygous for the 10th generation. The genomic integration of RNAiDENV in the mosquito genome was confirmed using Southern blot analysis using restriction enzymes ScaI or EcoRI (Fig. 1F,G).
Molecular analysis of synthetic miRshRNA expression and processing
A qPCR was performed to confirm the expression and processing of the selected DENV targeting sequences of the miRshRNAs (Fig. 2A), and the qPCR results of each miRshRNAs were normalized using the normalization method described in Yen et al., 2018, in comparison to host-specific mir-1174 (miRBase.org ref: MI0013471). The differential expression of miRshRNA was found in the midgut of mosquitoes relative to mir-1174 (One-way ANOVA, p < 0.05) (Fig. 2B), and the expression level values for miRK1.1, miRK2.6, miR14.5, and miR37.4 were 1.5026 ± 0.4212, 2.04411 ± 0.57553, 0.03844 ± 0.0155, and 0.00380 ± 0.00156, respectively. Gel image analyses showed the PCR products were in the correct size range of miRshRNA [70bp to 80bp; Fig. 2C)].
Figure 2.
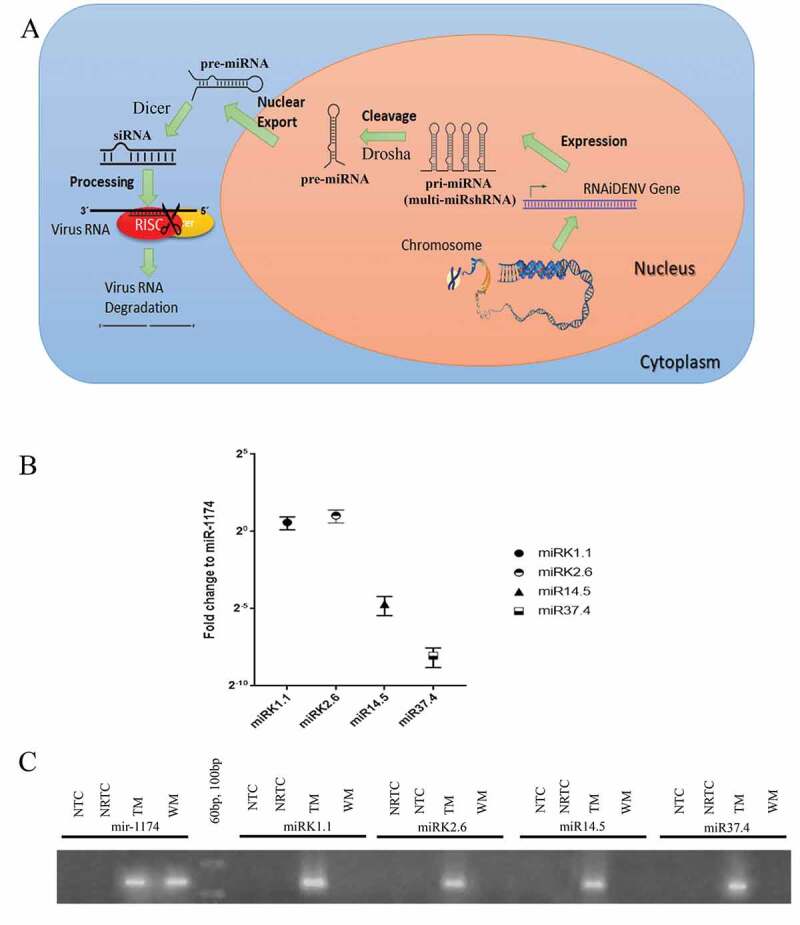
Biogenesis and expression analyses of multi-miRshRNA in transgenic Ae. aegypti. (A) Schematic diagram showing processing multi-miRshRNA of RNAiDENV to miRNA by RNAi pathway. (B) Relative expression levels of DENV target sequences in TM with respect to the reference, Ae. aegypti microRNA, mir-1174 at 1 dpi, (C) Gel image of miRshRNAs and mir-1174 expressions at 1 dpi in WM and TM. Three replicates were performed in analysing relative expression levels of DENV target sequences at 1 dpi. Bars display mean ± SEM (Standard Error Mean) (TM: transgenic mosquito, WM: wild mosquito, miRshRNA: microRNA based shRNA, multi-miRshRNA; multiple miRshRNA, pri-miRNA: primary miRNA, pre-miRNA: precursor miRNA, siRNA: small interfering RNA, Dicer and Drosha are enzymes involved in RNA interference mechanism, RISC: RNA-induced silencing complex, RNAiDENV: Designed gene with AeCPA/multi-miRshRNA/svA, NRTC: No reverse transcriptase control, NTC: No template control).
Dengue challenging experiments and dengue virus transmission
The challenging experiment was conducted by feeding mosquitoes with the DENV infected blood. The measurements of DENV infection, dissemination, and transmission rates in individual mosquitoes based on qPCR assay demonstrated that DENV2 and DENV4 have significantly reduced infection rates in hemizygous transgenic mosquitoes (TM) at 14 dpi, compared to wild mosquitoes (WM), Ae. aegypti (Fig. 3). Fisher’s exact test values were as follows: for infection rates – DENV2: WM: 82.67%, TM: 9.33%, p < 0.005; and DENV4: WM: 81.33%, TM: 14.66%, p < 0.005; for dissemination rates – DENV2: WM: 50.67%, TM: 9.33%, p < 0.005; and DENV4: WM: 69.33%, TM: 6.62%, p < 0.005; and for transmission rates – DENV2: WM: 41.33% TM: 6.67%, p < 0.005; and DENV4: WM: 37.33% TM: 5.33%, p < 0.005. To analyse this further, immunohistochemical analyses were performed using anti-NS1 antibody and FITC labelled secondary antibody (Fig. 4) and in this assay, the DENV2 and DENV4 fed TM midgut tissues showed a reduced fluorescent intensity compared to WM, confirming that the RNAiDENV is interrupting the replication of DENV2 and DENV4.
Figure 3.
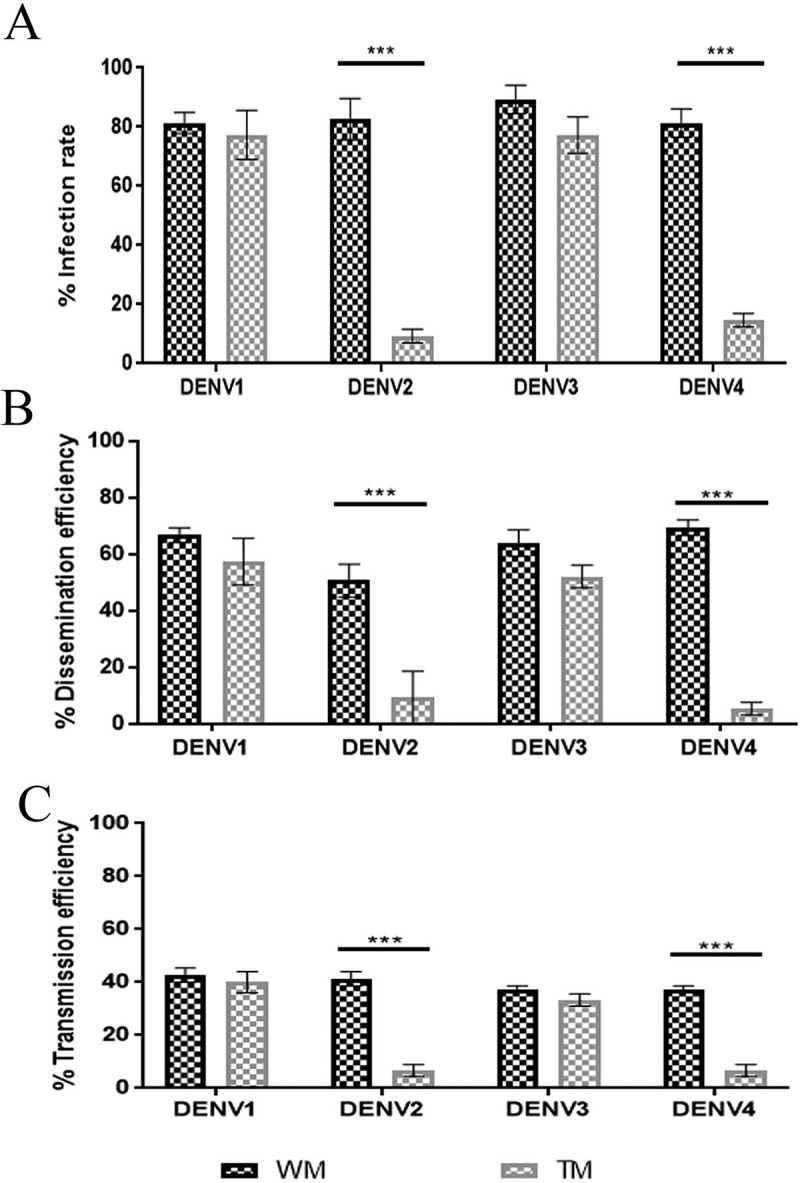
DENV serotype challenging analysis in transgenic Ae. aegypti. (A) Infection rate, (B) Dissemination rate, (C) Transmission rate. Mosquitoes were challenged with DENV1, DENV2, DENV3, and DENV4 at titres of 107 ffu/mL. Samples were collected and titrated at 14 dpi. The infection rate was defined as the number of positive body samples among tested ones; dissemination efficiency refers to the number positive in the midgut samples (i.e., successful viral dissemination after development of virus in the midgut barrier) among tested ones; transmission rate was defined as the number of positive saliva samples (i.e., successful transmission) among tested ones. DENV challenging study was carried out with three replicates and each with 25 mosquitoes (3 x 25). Bars represent mean ± SEM (Standard Error Mean). Fisher’s exact test was performed to analyse the samples. Significant differences at P < 0.05 are indicated by ***. (ffu: focus forming unit, TM: transgenic mosquito, WM: wild mosquito, DENV: dengue virus, DENVn: dengue virus serotype n).
Figure 4.
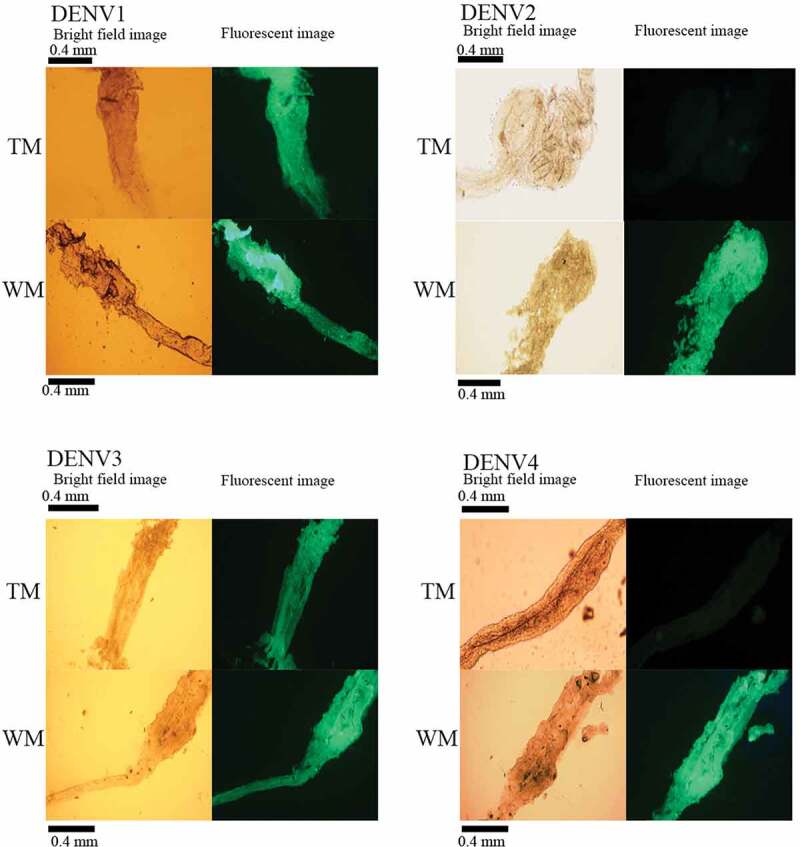
Immunohistochemical analyses of NS1 antigen expression in the midguts of TM and WM by FITC labelled secondary antibody following challenge with DENV serotypes. (A) DENV1 infected (B) DENV2 infected (C) DENV3 infected (D) DENV4 infected. Fluorescence was visualized using Olympus BX53 with the FITC filter. Magnification is 20X and bright-field images and their corresponding fluorescence images are shown. (TM: transgenic mosquito, WM: wild mosquito, DENV: dengue virus, DENVn: dengue virus serotype n, FITC: Fluorescein isothiocyanate).
Impact of the transgene construct on mosquito fitness in laboratory trials
To determine whether the transgene has any significant effect on the fitness, the hemizygous TM line, SL1161 was compared with the WM Ae. aegypti colony in a laboratory trial using fitness parameters, such as oviposition, fecundity/fertility, larval life span, and adult male and female life span (Fig. 5). Female oviposition for TM was 50.98 ± 1.80 (n = 168), and this was significantly lower than that of WM, 77.62 ± 1.988 (n = 188) (unpaired t-test: p < 10−4). Similarly, the average hatchability of eggs/fertility for TM was 0.55 ± 0.02, (n = 168) and this was lower than that of WM at 0.84 ± 0.01 (n = 188) (unpaired t-test: p < 10−4). The average life span of adult male TM was 31.83 ± 8.94 days and this was significantly shorter than that of WM, 39.71 ± 10.04 days. The average life span of female TM was 37.02 ± 13.46 days and this was shorter than that of WM 45.54 ± 12.70 days. Also, survival curves of adult male and female TM were significantly different from those of WM (Log-rank (Mantel-Cox) test: p < 10−4) (Fig. 6).
Figure 5.
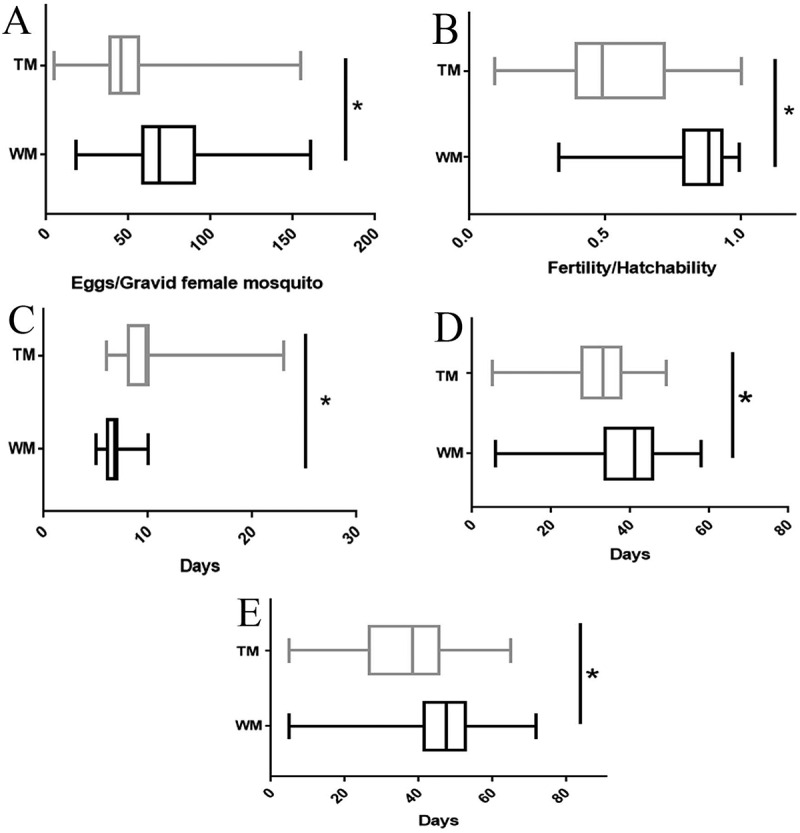
Fitness parameter measurements of TM in comparison to WM. (A) Oviposition, (B) Fertility, (C) Larval life span, (D) Adult male life span and (E) Adult female life span. Oviposition defined as the number of eggs per blood-fed female mosquitoes and female mosquitoes without any eggs laid was excluded from results; fecundity as the number of L1 larvae/number of eggs; larval life span as the number of days to become pupae from larvae; adult life span as the number of days to the death of the mosquitoes. Three replicates were performed for each experiment. Bars of A, B, C, D, and E represent the maximum value to the minimum value of the results of TM or WM. The unpaired t-test was performed to analyse the parameters between WM and TM. Significant differences at P < 0.05 are indicated by *. (TM: transgenic mosquito, WM: wild mosquito).
Figure 6.

Survival curves of WM and TM. (A) Males. (B) Females. Survival curves were compared between WM and TM. Approximately, 200 mosquitoes of TM and WM were used in this experiment. (TM: transgenic mosquito, WM: wild mosquito).
Discussion
We designed an RNA interference (RNAi) gene, RNAiDENV containing multiple microRNA-based short hairpin RNA (multi-miRshRNA). These multi-miRshRNAs of RNAiDENV are transcribed by RNA polymerase II into a polycistronic transcript containing pri-miRNAs and are subjected to post-transcriptional processing by Drosha to form pre-miRNA in cells prior to transportation from the nucleus. In the cytoplasm, pre-miRNAs are further processed by the Dicer into short double-stranded RNA fragments, small interfering RNA. RNA-induced silencing complex (RISC) activated by Dicer for RNA interference together with small interfering RNA. The catalytic component of RISC degrades or silences the expression of target RNAs [22,23]. RNAiDENV was designed in a way to trigger the endogenous RNAi pathway mentioned above against the replication of dengue virus (DENV) in Aedes aegypti. A similar approach has been implemented in a study using a RNAi molecule to turn off the expression of green fluorescent protein mRNA in 293 T cells, showing that the knockdown efficiency of the latter is greater when using a triple hairpin construct than when using a single-hairpin or double-hairpin of artificially modified miR-30 [24]. Further, another study using microRNA-like short hairpin RNAs, kept under the control of RNA polymerase II promoter, has shown the robust depletion of expression of multiple genes such as arrestin 2, G beta 2 and G-protein coupled receptor kinase 2 in the RAW264.7 murine macrophage cell line [25]. Therefore, the foregoing study is another step forward, whereby combining the approaches mentioned above in a living system, having single promoter-driven, multiple target miRNAs gives rise to multiple RNA interference for the DENV virus serotypes in transgenic Ae. aegypti.
DENV has four different serotypes and targeting one serotype will result in unusual dominance of the rest of serotypes over that serotype, therefore, simultaneous targeting of multiple serotypes is vital [26]. This premise is implemented in our study by designing multiple DENV serotypes targeting multi-miRshRNA, and making each miRshRNA in the design target two or more serotypes, or vice-versa (Table 1) to avoid the development of escape mutants and to reduce the size of the RNAi triggering effector molecule to increase the transformation efficiency. The aim of the design was to keep a smaller number of secondary structural isoforms for the multi-miRshRNA to have structural stability by reducing the size of the RNA molecule, using a multiple DENV serotype targeting shRNA and ensuring only two structural isoforms for the multi-miRshRNA in mosquito cells having ΔGs of −247.38 kcal/mol and −233.60 kcal/mol for the stable and other isoform, respectively. Selecting the most effective DENV target sequences are critical in designing dengue resistant multi-miRshRNA. Previous studies have suggested DENV capsid, prM, NS5, and NS4B regions as the critical targets to activate RNAi mechanism [9,27], and on that basis, DENV target sequences (Fig. 1A, Table 1) were selected and analysed for mismatch tolerance/off-target effect with respect to the mosquito transcriptome, using BLAST search tool of NCBI. The maximum tolerance was set as more than 17 bp contiguous base matches complementary to matching sequences of mosquito transcriptome. The mir-1175 (Ae. aegypti miRNA-1175, miRBase.org ref: MI0013470) of Ae. aegypi has been recognized as the most suitable template for designing miRshRNAs as it has attributes of host-specific processing and blood-meal induced expression in the midgut [28]. Therefore in designing the miRshRNA molecule, mir-1175 was used as a template while leaving its pre-microRNA loop and tail sequences and replacing its mature microRNA sequence with the selected target sequence, as demonstrated in Sun et al., 2006 (Fig. 1B). Similar approaches have recently been adopted in designing miRshRNA-like elements using Drosophila specific miRNAs by Yen et al., 2018 and Buckman et al, 2019, to achieve the resistance in Ae. aegypti to arboviruses of DENV3, Chikungunya (CHIKV) and Zika virus.
To integrate the designed RNAiDENV containg multi-miRshRNA to the Ae. aegypti Sri Lankan strain, the piggyBac transposable element containing cargo plasmid was used, as the latter element is host factor-independent and highly efficient in transposition in a wide range of organisms such as yeast, insects, planarian, malaria parasite, and mammals [29,30], and other strains of Ae. aegypti [31]. The observation of reporter gene expression in compound eyes of larval and adult stages of transgenic mosquitoes (Fig. 1E) indicated that piggyBac is capable of integrating a transgene construct into the Ae. aegypti Sri Lankan strain. The integration of the transgene construct into the genome of the Ae. aegypti Sri Lankan strain was proven further by nested PCR (data not shown) and Southern blot hybridization (Fig. 1F,G). Furthermore, the observation of expression of individual miRshRNA of RNAiDENV in TM in qPCR (Fig. 2B,C) with shRNA-specific primers (Table 2) indicated that the individual miRshRNA of RNAiDENV is processed in mosquitoes to synthesize siRNA, and it was also noted that, in spite of these, the differential expression of the miRshRNA was done under a single promoter, indicating a positional effect of miRshRNA expression. Similar observations have been reported when synthetic microRNAs under the control of a single promoter was used to knock down DENV3 and Chikungunya viruses [16].
Table 2.
Chosen target sequence detection primers.
| No | Primer Name | Primer Sequence | Tm | GC% | Remark |
|---|---|---|---|---|---|
| 1 | miRK1.1_1_A | AAGAGACACAGGATCTCTGG | 62.3ºC | 52.40% | Forward Primer |
| 2 | miRK2.6_1_A | AAAGCATCAGAGATCTGCTCTC | 61.3ºC | 45.50% | Forward Primer |
| 3 | miR14.5_1_A | AAGCTTTGCATTTGC TCCAG | 61.1ºC | 45.00% | Forward Primer |
| 4 | miR37.4_1_A | AAGTCAGCATATTGATAGACGG | 59.3ºC | 40.90% | Forward Primer |
| 5 | Reverse Primer | miScript Universal Primer | - | - | Reverse Primer |
Tm- Melting temperature, GC%- Guanine and Cytosine percentage.
Dengue challenging study showed that the multi-miRshRNA was able to knock down the DENV2 and DENV4 but was not that effective for DENV1 and DENV3. A previous study, based on an adenoviral delivery of DENV target sequences embedded in shRNAs, named sh-3d (miRK1.1) and sh-5b (miRK2.6), to the HEK 293 cell line infected with dengue viruses, gave a similar observation, where a strong reduction of DENV2 and DENV4 compared to DENV1 and DENV3 was shown. The reduction of DENV2 by 92.6% and DENV4 by 80.3%, (DENV1 by 16.6% and DENV3 by 0%) by miRK1.1, and DENV2 by 48.2% and DENV4 by 70.8% (DENV1 by 51.3%, and DENV3 by 63.8%) by miRK2.6 was reported in this study [18]. Our study too elicited a similar trend of reduction of infection rate of DENV2 by 90.67% and DENV4 by 85.34% in TM indicating targeting efficiency of DENV2 and DENV4 by miRshRNAs. The ineffectiveness of miRshRNAs to target DENV1 and DENV3 is perhaps due to the association with ribonucleic viral proteins with the target sequences of DENV [32] to prevent the access of the RISC complex to the latter sequences, although strict sequence matching is seen between the sequences used in the design and target sequences of DENV in the bioinformatic analyses; or it may be due to some of miRshRNAs (miR37.4 and miR14.5) not being expressed up to the physiologically relevant level with respect to that of mir-1174 in Ae. aegypti. Further, virus escape to RNAi may be due to the acquisition of mutations in the targeted sequences, as it is required to have at least 80% sequence identity to the viral RNA target to activate RNAi pathways [33,34]. Adelman and co-workers [35] demonstrated dsRNA sequence mediated RNAi resistance to DENV2 in Ae. aegypti mosquitoes while Yen and co-workers (2018) showed partial resistance to DENV3 and Chikungunya virus (CHIKV) in transgenic Ae. aegypti through the induction of the expression of synthetic miRNAs [16]. But some of the drawbacks of their studies was the inability to target multiple serotypes of DENV simultaneously in a mosquito, and the use of larger RNAi gene constructs (dsRNA and synthetic miRNA) for genetic transformation in mosquitoes. However, in the foregoing study, we showed that the smaller sequence (approximately 400 bp) can block DENV transmission in transgenic Ae. aegypti and that these mosquitoes may be useful in epidemic situations to block disease transmission. However, release of the transgenic mosquito into the environment requires several other precautions and assessments. Laboratory fitness is one of the preliminary assessments that have to be carried out and thus, we tested TM oviposition, fecundity and, larval and adult life span in comparison to WM, and found comparative reduction of oviposition, fecundity, adult life span, in spite of longer life span for TM larvae. These results are common to transgenic mosquitoes, and the same was revealed by Catteruccia et al. [36], where the reduction of fitness of four different TM lines expressing fluorescent reporter proteins under the control of actin promoter, compared to WM, was reported.
In conclusion, the strategy detailed in this study can be used to develop not only a transgenic Ae. aegypti with resistance to all known DENV serotypes but also can be used to engineer an Aedes mosquito resistant to other arboviruses like Yellow fever virus, Japanese encephalitis virus, West Nile virus and CHIKV. To our knowledge, this is the first study to demonstrate the resistance to multiple DENV serotypes in transgenic Ae. aegypti together with analysing the effects of the transgene on the mosquito fitness. This will greatly contribute towards the development of transgenic mosquitoes with the required level of fitness and potentially benefit dengue control programmes.
Materials and methods
Ethics statement
Ethical clearance for the experimental procedures was obtained from the Ethics Review Committee – University of Kelaniya, application number P/149/08/2018. Dengue challenging, mosquito rearing, and microinjection procedures were performed in the Arthropod Containment Level-2 (ACL-2) facility at Molecular Medicine Unit, as described in ethical statements of Ethics Review Committee of the Faculty of Medicine, University of Kelaniya, Sri Lanka
Identification of target sequences for DENV serotypes
DENV genomic sequences of Sri Lankan origin, DENV serotype 1 (GenBank access numbers JN054256.1, JN054255.1, HQ891316.1, HQ891315.1, HQ891314.1, HQ891313.1, KJ726664.1, KJ726662.1, KJ726663.1, KJ468234.1) and 3 (GenBank access numbers KF955476.1, KF955474.1, NC_001475.2, FJ882574.1, FJ882573.1, FJ882572.1, FJ882571.1, JQ411814.1, GQ199889.1, GQ252674.1, GQ199888.1, GQ199887.1, AY099336.1) were retrieved from GenBank and they were multiply aligned using BioEdit v7.1.11, and consensus sequences were obtained from the aligned sequences. Two sequences (~21 bp) to target the DENV type 1 and 3 (Table 1) were selected from the DENV non-structural protein-coding sequence (NS5) [37] and capsid consensus region of structural protein-coding sequence [27,38]. Target sequences of miRK1.1 and miRK2.6 (Table 1) were chosen from a previous study from the putative serine target sites of 5ʹ upstream and 3ʹ downstream untranslated regions of DENV [18]. Each serotype was targeted with 2 or more DENV targeting sequences.
Off-targeting effect on Ae. aegypti transcriptome by selected target sequences
All four DENV targeting sequences were BLAST searched against Aedes aegypti sequences deposited in the GenBank and RefSeq databases of the National Centre for Biotechnology Information (NCBI,http://www.ncbi.nlm.nih.gov/BLAST/) using ‘search for short nearly exact matches’ mode. These searches were performed to find any significant homology (> 17 contiguous nucleotides of identity) with sequences of the Ae. aegypti.
Designing multi-miRshRNA transcript and prediction of secondary structure
MicroRNA-based shRNA (miRshRNA) sequences were designed from the microRNA loop and tail sequences of Ae. aegypi miR-1175 (miRBase.org ref: MI0013470) by replacing the mature microRNA sequence of miR-1175 with the selected DENV target sequence and the complementary strand of the selected DENV target sequence given in Fig. 1A. The resulting four miRshRNA sequences and a signalling sequence were connected together using appropriate restriction enzyme sites to obtain a multiple miRshRNA (multi-miRshRNA) nucleotide sequence. The UNAfold programme [39,40] of the mfold web server was used to predict and optimize the secondary structure for multi-miRshRNA nucleotide sequence. The thermodynamic interactions and RNA folding patterns of each shRNA of miRshRNA were analysed from the mfold RNA folding tool. The energy dot-plot and secondary structures were analysed to ensure the formation of four miRshRNA structures of multi miRshRNA transcript in bioinformatics analysis.
Designing transgene RNAiDENV
RNAiDENV was designed by incorporating the Ae. aegypti carboxypeptidase A promoter (AeCPA), multi-miRshRNA and Simian virus 40 terminator sequence (svA). The sequences of RNAiDENV DNA (GenBank accession no: MK861918) are given in Figure S3. The transgene construct was prepared by cloning RNAiDENV into the AscI and MluI restriction site of the cargo plasmid (pBac-3xp3DsRED, donated from ICB II, USP, Brazil). The order of elements contained in the transgene cassette are piggyBac left sequence, reporter gene, RNAiDENV and piggyBac right sequence (Fig. 1D).
Mosquito rearing
For general rearing, Sri Lankan wild Ae. aegypti strain was maintained at 28°C, 72-80% relative humidity, under a 14–15 hr light and 9–10 hr dark cycle, in the ACL-2 facility at the Molecular Medicine Unit, Faculty of Medicine, University of Kelaniya, Sri Lanka. A feeding solution [glucose 10% (W/V) and Vitamin B{Polybion, Merk Ltd, India} solution 2% (V/V)] was used to feed adult mosquitoes while cattle blood was used to feed Ae. aegypti to lay eggs.
Transgene construct transformation and analysis
Preblastoderm-stage embryos were collected and microinjected with the microinjection solution containing pBacRNAiDENV (Fig. 1D) plasmid (0.5 µg/µL) and pHelper (0.3 µg/µL) using TransferMan® 4 r (Eppendorf, Germany) and FemtoJet® 4i (Eppendorf, Germany) system. An optimized injection pressure range of 450–600 hPa, a back-pressure range of 150–200 hPa, and time 0.8–1.2 s was used for microinjections (methodology described in supplementary data). Approximately 5-10% of egg volume was microinjected to eggs by comparing the egg size. pBacRNAiDENV and pHelper (Plasmid expressing piggyBac transposase) plasmid with MIB injected embryos were heat-shocked at 39°C after 16 hours post-injection for 1 hour, to induce the transposase enzyme which catalyzes the movement of the construct to the genome, and then they were kept at 28°C, 82% humidity for about 5 days further development. The embryos were allowed to develop till the adult stage (G0 generation) and each surviving G0 male mosquito was out-crossed to five wild females and each surviving G0 female mosquito was outcrossed with two wild males. Transgenic lines were maintained as hemizygous by out-crossing transgenic individuals at every generation with wild-type mosquitoes. Progeny of these crosses (G1) was screened for DsRED protein (Red fluorescent protein) expression in their eyes using Olympus BX53 fluorescence microscope to recover transgenic mosquitoes (TM). The strong miRshRNA expressing TM line, TRSL1161 was selected for further studies. This transgenic line was maintained as hemizygous (until the 10th generation) by out-crossing transgenic individuals at every generation with wild-type mosquitoes. Expression of miRshRNAs was carried out with 5th and 7th generations of TM line and dengue challenging assays and laboratory fitness measurements were carried out using the 7th generation of the TM line, comparing with wild mosquitoes (WM).
Southern blot analysis
Genomic DNA of the TM and WM was extracted using a GenoSpinRTM Total DNA Extraction Kit (CEYGEN Biotech, Colombo, Sri Lanka) according to the manufacturer’s instructions. A total of 20 µg of extracted genomic DNA, digested with restriction enzymes EcoRI or ScaI, was separated on 0.8% agarose gel overnight. Separated genomic DNA was then transferred onto a nylon membrane by upward capillary transfer and immobilized by UV irradiation. Hybridization was performed using a biotin-labelled double-stranded probe comprising the svA and piggyBac right sequence. The nylon membrane was prehybridized for 5 h at 65 ° C in a prehybridization buffer containing 6× SSC (0.9 M NaCl/0.09 M trisodium citrate [pH 7.0]), 0.5% SDS, 5× Denhardt’s (0.1% Ficol 400, 0.1% polyvinylpyrrolidone, 0.1% BSA) and 100 µg/ml denatured salmon sperm DNA (Cat#D1626 Sigma,USA). Hybridization with the biotin-labelled probe was performed overnight at 50 ° C in a rotating tube in a hybridization buffer (6 × SSC, 0.5% SDS, 5× Denhardt’s, 100 µg/ml denatured salmon sperm DNA, and denatured hybridization probe concentration final to 50 ng/mL). After hybridization, the filter was washed successively in 2 x SSC containing 0.1% sodium dodecyl sulphate (SDS) for 5 min at room temperature, 2 x SSC containing 0.1% SDS for 15 min at 65 ° C, and 0.2 x SSC containing 0.1% SDS for 15 min at 65 ° C. The membrane was washed with 2× SSC for 10 min at room temperature. The membrane was incubated for 30 min at room temperature in 1/1000 AP-conjugated anti-mouse Ab (Roche) followed by colour development using NBT/BCIP (Cat# 69,219, Sigma, USA).
Detection of siRNAs derived from the midgut tissue of female mosquitoes
The total microRNA of TM and WM female mosquitoes fed with cattle blood and starved for 24–48 hr, was extracted using a miRNeasy Mini Kit (Qiagen, Valencia, CA, USA) [41] and cDNA was prepared from the extracted microRNA pools of 5 mosquitoes using miScript reverse transcription kit (Qiagen). Mature miRNA assay was performed using 5x Hot FirePol® Evagreen® qPCR Mix Plus (Solis Biodyne, Estonia) with miScript universal primer (U6B) and the DENV target-specific primers (Table 2) according to the manufacturer’s instructions [42] using the optimized temperature program given below. The qPCR mixtures were incubated at 95°C for 12 min to activate the HotStart DNA Taq polymerase, followed by 40 cycles of 95°C for 15 s and 58.5°C for 60 s, monitored by melt curve analysis. Ct values >35 were excluded from the analyses and Ae. aegypti specific microRNA mir-1174 (miRBase.org ref: MI0013471) was used as the internal control (house-keeping control) to normalize the DENV target sequences expression using as described by Yen et al., 2018 in transgenic Ae. aegypti. The resulting qPCR products were analysed on a 3% agarose gel using gel electrophoresis.
Dengue infection and dissemination analysis of transgenic mosquitoes
TM and WM mosquitoes were infected with a blood meal containing 107 ffu/mL of DENV using the Glytube method [43], and maintained for 14 days under a sealed container. Prior to mosquito infections, DENV infected cell culture samples obtained from the Department of Microbiology, University of Sri Jayewardenepura, Sri Lanka, were reassessed for serotypes using RT-PCR [44]. Viral RNA was extracted using QIAamp viral RNA Mini Kit (Qiagen, Germany) after 14 days post-infection (dpi) and cDNA synthesis of each sample was performed using FIREScript RT cDNA synthesis Mix (Solis Biodyne, Estonia). Viral RNA expression was analysed by RT-qPCR using 5x Hot FirePol® Evagreen® qPCR Mix Plus (Solis Biodyne, Estonia) according to the manufacturer’s instructions [45] and Ct values >35 were excluded from the analyses. Positives samples were identified using melt curve analysis and agarose gel electrophoresis. The infection rate refers to the proportion of mosquitoes with infected body among engorged mosquitoes. The dissemination rate corresponds to the proportion of mosquitoes with the infected heads among mosquitoes with infected bodies. (ffu: focus forming unit; RT-qPCR: Reverse transcription quantitative PCR; RT-PCR: Reverse transcription PCR)
Transmission assay
Saliva from individual females orally exposed to DENV was collected at 14 dpi for virus detection. TM and WM were anesthetized by keeping on ice, transferred to a glass petri dish on ice and then the legs and wings were removed with a fine syringe needle and sterile forceps. A Plastic slide with a strip of modelling clay was used to support micropipette tips filled with 10 μL of Leibovitz’s L-15 medium (Cat#F7524 Sigma-Aldrich, USA) with 2% foetal bovine serum albumin (Cat#L4386 Sigma-Aldrich, USA) and the proboscis of each live mosquito was inserted into the tip. Insects were allowed to salivate for 45 minutes and the total volume was collected into the 1.5 mL microtube. After salivation, mosquito saliva was injected intrathoracically into 25 female mosquitoes, 5–7 days old, from a 24 hr post-blood-fed wild type colony [45–48]. Viral RNAs were extracted using QIAamp viral RNA Mini Kit (Qiagen, Germany) after 14 dpi, and cDNA synthesis of each sample was performed using FIREScript RT cDNA synthesis mix (Solis Biodyne, Estonia). Viral RNA expression analysis was performed with qPCR using 5x Hot FirePol® Evagreen® qPCR Mix Plus (Solis Biodyne, Estonia) according to the manufacturer’s instructions, and Ct values >35 were excluded from the analyses. Positives samples were identified using melt curve analysis and agarose gel electrophoresis. The transmission rate represents the proportion of mosquitoes with infectious saliva among mosquitoes examined.
DENV NS1 antigen expression analysis in the midgut
DENV infected TM and WM were dissected to peel out midgut tissues and fixed in microscopic glass slides, and the dissected midguts were fixed in 4% (v/v) paraformaldehyde in PBS. Immunohistochemical staining (IHC) was performed by using the rabbit anti-DENV NS1 antibody (Sigma, USA cat no. SAB2700186) as the primary antibody and FITC labelled goat anti-rabbit antibody (Abcam, USA cat no. ab6717) as the secondary antibody, to detect viral antigen [49,50] using the protocol described in Costa et al. (1980). Fluorescence produced by bound FITC labelled antibodies was visualized using the Olympus BX53 fluorescence microscope [51].
Fitness factor assessment
Fitness tests were assessed in hemizygous transgenic mosquitoes with respect to their wild type siblings, and standard procedures were used to treat them equally in the insectary facility and ensure that differences in fitness traits measurements did not result from handling or environmental conditions. One hundred larvae were checked daily until pupation and adult emergence, and mosquitoes were maintained in the Arthropod Containment Level 2 condition, with larval and pupal diet described elsewhere, for adult lifespan analysis. The parameters of fecundity were the number of eggs laid per female, and egg hatchability, the number of larvae emerged/number of eggs. To synchronize larval life span, more than 200 eggs were hatched and 200 L1 larvae were collected, and the larval life span (the number of days taken to become pupae) was obtained. Adult life span was synchronized by selecting a batch of pupae that emerged on the same day and adult life span was calculated by counting dead mosquitoes with days. Larval and adult lifespan tests were performed in three replicates.
Data analyses
Data analysis was conducted using the GraphPad Prism5 software package (Version 5.00) for Windows (San Diego, California, USA) and confidence intervals of 95% were defined for all analyses. Oneway ANOVA (Analysis of variance) was used to compare means of relative expression levels of DENV target sequences. Fisher’s test was performed to analyse the infection rates, dissemination rates and transmission rate of WM and TM. The unpaired t-test was applied to compare fecundity, oviposition and larval life span of each WM and TM. Mosquito life tables were analysed using the Log-rank (Mantel-Cox) test [52] and the average life span was calculated for both sexes of TM and WM using Microsoft Excel.
Supplementary Material
Acknowledgments
We thank Dr. D. Carvalho (Insect Pest Control, IAEA) for generously donating piggyBac plasmids, Dr. H. Corrêa de Araújo Gomes (ICB, University of Sao Paulo, Brazil) for providing microinjection training, Mr. Channa Aluwihare (University of Maryland, USA) for helping to identify transgenic mosquitoes, Prof. N. Malavige (University of Sri Jayewardenepura, Sri Lanka) for donating DENV samples and cultures for experiments, Mr.K. Muneeswaran (University of Colombo, Sri Lanka) for helping in bioinformatic analyses, Dr. N.V. Chandrasekharan (University of Colombo, Sri Lanka) and Prof. W. Abeyewickreme (Kothalawala Defense University, Sri Lanka) for technical and administrative support.
Funding Statement
We acknowledge the National Research Council [TO 14-04] for funding and the International Atomic Energy Agency [IAEA SRL5/047] for technical co-operation;National Research Council Sri Lanka [TO 14-04].
Disclosure Statement
No potential conflict of interest was reported by the authors.
Supplementary Material
Supplemental data for this article can be accessed here.
References
- [1].Blair CD, Adelman ZN, Olson KE.. Molecular strategies for interrupting arthropod-borne virus transmission by mosquitoes. Clin Microbiol Rev. 2000;13(4):651–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Malavige GN, Fernando S, Fernando DJ, et al. Dengue viral infections. Postgrad Med J. 2004;80:588–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Idrees S, Ashfaq UA. RNAi: antiviral therapy against dengue virus. Asian Pac J Trop Biomed. 2013;3(3):232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Malavige GN, Fernando N, Ogg G. Pathogenesis of dengue viral infections. Sri Lankan J Infect Dis. 2011;1(1):2. [Google Scholar]
- [5].Labbé GMC, Scaife S, Morgan SA, et al. Female-specific flightless (FsRIDL) phenotype for control of aedes albopictus. PLoS Negl Trop Dis. 2012;6(7):e1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Qsim M, Ashfaq UA, Yousaf MZ, et al. Genetically modified aedes aegypti to control dengue: a review. Crit Rev Eukaryot Gene Expr. 2017;27(4):331–340. [DOI] [PubMed] [Google Scholar]
- [7].Monath TP. Treatment of yellow fever. Antiviral Res. 2008;78:116–124. [DOI] [PubMed] [Google Scholar]
- [8].Caglioti C, Lalle E, Castilletti C, et al. Chikungunya virus infection: an overview. New Microbiol. 2013;36:211–227. [PubMed] [Google Scholar]
- [9].Travanty EA, Adelman ZN, Franz AWE, et al. Using RNA interference to develop dengue virus resistance in genetically modified aedes aegypti. Insect Biochem Mol Biol. 2004;34(7):607–613. [DOI] [PubMed] [Google Scholar]
- [10].Carvalho DO, Nimmo D, Naish N, et al. Mass production of genetically modified aedes aegypti for field releases in Brazil. J Vis Exp. 2014;83. DOI: 10.3791/3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wilke ABB, Beier JC, Benelli G. Transgenic mosquitoes – fact or fiction? Trends Parasitol. 2018;34:456–465. [DOI] [PubMed] [Google Scholar]
- [12].Catteruccia F, Crisanti A, Wimmer EA. Transgenic technologies to induce sterility. Malar J. 2009;8(SUPPL. 2). DOI: 10.1186/1475-2875-8-S2-S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Johnson R, Guigó R. The RIDL hypothesis: transposable elements as functional domains of long noncoding RNAs. Rna. 2014;20(7):959–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Armas-Tizapantzi A, Montiel-González AM. RNAi silencing: a tool for functional genomics research on fungi. Fungal Biol Rev. 2016;30:91–100. [Google Scholar]
- [15].Weinheimer I, Jiu Y, Rajamäki ML, et al. Suppression of RNAi by DsRNA-degrading RNaseIII enzymes of viruses in animals and plants. PLoS Pathog. 2015;11(3):1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yen PS, James A, Li JC, et al. Synthetic MiRNAs induce dual arboviral-resistance phenotypes in the vector mosquito aedes aegypti. Commun Biol. 2018;1(1). DOI: 10.1038/s42003-017-0011-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sánchez-Vargas I, Scott JC, Poole-Smith BK, et al. Dengue virus type 2 infections of aedes aegypti are modulated by the mosquito’s RNA interference pathway. PLoS Pathog. 2009;5(2):e1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Korrapati AB, Swaminathan G, Singh A, et al. Adenovirus delivered short hairpin RNA targeting a conserved site in the 5′ non-translated region inhibits all four serotypes of dengue viruses. PLoS Negl Trop Dis. 2012;6(7):e1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhang T, Cheng T, Wei L, et al. Efficient inhibition of HIV-1 replication by an artificial polycistronic MiRNA construct. Virol J. 2012;9(1):118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Buchman A, Gamez S, Li M, et al. Engineered resistance to zika virus in transgenic aedes aegypti expressing a polycistronic cluster of synthetic small RNAs. Proc Natl Acad Sci U S A. 2019;116(9):3656–3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Schetelig MF, Handler AM. A functional comparison of the 3xp3 promoter by recombinase-mediated cassette exchange in drosophila and a tephritid fly, anastrepha suspensa. G3 Genes, Genomes, Genet. 2013;3(4):687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wahid F, Shehzad A, Khan T, et al. MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim Biophys Acta Mol Cell Res. 2010;1803:1231–1243. [DOI] [PubMed] [Google Scholar]
- [23].Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42(1):217–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sun D, Melegari M, Sridhar S, et al. Multi-MiRNA hairpin method that improves gene knockdown efficiency and provides linked multi-gene knockdown. Biotechniques. 2006;41(1):59–63. [DOI] [PubMed] [Google Scholar]
- [25].Zhu X, Santat LA, Chang MS, et al. A versatile approach to multiple gene RNA interference using microRNA-based short hairpin RNAs. BMC Mol Biol. 2007;8:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Stove V, Smits K, Naessens E, et al. Multiple gene knock-down by a single lentiviral vector expressing an array of short hairpin RNAs. Electron J Biotechnol. 2006;9(5):572–579. [Google Scholar]
- [27].Haasnoot PCJ, Cupac D, Berkhout B. Inhibition of virus replication by RNA interference. J Biomed Sci. 2003;10:607–616. [DOI] [PubMed] [Google Scholar]
- [28].Li S, Mead EA, Liang S, et al. Direct sequencing and expression analysis of a large number of MiRNAs in aedes aegypti and a multi-species survey of novel mosquito MiRNAs. BMC Genomics. 2009;10:581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhao S, Jiang E, Chen S, et al. PiggyBac transposon vectors: the tools of the human gene encoding. Transl Lung Cancer Res. 2016;120–125. DOI: 10.3978/j..2218-6751.2016.01.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Woltjen K, Michael IP, Mohseni P, et al. PiggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458(7239):766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kistler KE, Vosshall LB, Matthews BJ. Genome engineering with CRISPR-Cas9 in the mosquito aedes aegypti. Cell Rep. 2015;11(1):51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Haasnoot J, Westerhout EM, Berkhout B. RNA interference against viruses: strike and counterstrike. Nat Biotechnol. 2007;25:1435–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Presloid JB, Novella IS. RNA viruses and RNAi: quasispecies implications for viral escape. Viruses. 2015;7:3226–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Birmingham A, Anderson EM, Reynolds A, et al. 3ʹ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006;3(3):199–204. [DOI] [PubMed] [Google Scholar]
- [35].Adelman ZN, Sanchez-Vargas I, Travanty EA, et al. RNA silencing of dengue virus type 2 replication in transformed C6/36 mosquito cells transcribing an inverted-repeat RNA derived from the virus genome. J Virol. 2002;76(24):12925–12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Catteruccia F, Godfray JCH, Crisanti A. Impact of genetic manipulation on the fitness of anopheles stephensi mosquitoes. Science. 2003;299(5610):1225–1227. [DOI] [PubMed] [Google Scholar]
- [37].Villegas PM, Ortega E, Villa-Tanaca L, et al. Inhibition of dengue virus infection by small interfering RNAs That target highly conserved sequences in the NS4B or NS5 coding regions. Arch Virol. 2018;163(5):1331–1335. [DOI] [PubMed] [Google Scholar]
- [38].Haasnoot J, Berkhout B. RNA interference: its use as antiviral therapy. Handb Exp Pharmacol. 2006;173:117–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol. 2008;453:3–31. [DOI] [PubMed] [Google Scholar]
- [40].Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ueno K, Hirata H, Shahryari V, et al. Tumour suppressor MicroRNA-584 directly targets oncogene rock-1 and decreases invasion ability in human clear cell renal cell carcinoma. Br J Cancer. 2011;104(2):308–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Son DJ, Kumar S, Takabe W, et al. The atypical mechanosensitive MicroRNA-712 derived from pre-ribosomal RNA induces endothelial inflammation and atherosclerosis. Nat Commun. 2013;4. DOI: 10.1038/ncomms4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Costa-da-Silva AL, Navarrete FR, Salvador FS, et al. Glytube: a conical tube and parafilm M-based method as a simplified device to artificially blood-feed the dengue vector mosquito, aedes aegypti. PLoS One. 2013;8(1):e53816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lanciotti RS, Calisher CH, Gubler DJ, et al. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30(3):545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Costa-Da-Silva AL, Ioshino RS, De Araújo HRC, et al. Laboratory strains of aedes aegypti are competent to Brazilian zika virus. PLoS One. 2017;12(2). DOI: 10.1371/journal.pone.0171951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Vazeille M, Mousson L, Martin E, et al. Orally co-infected aedes albopictus from La Reunion Island, Indian Ocean, can deliver both dengue and chikungunya infectious viral particles in their saliva. PLoS Negl Trop Dis. 2010;4(6):e706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Rosen L, Gubler D. The use of mosquitos to detect and propagate dengue viruses. Am J Trop Med Hyg. 1974;23(6):1153–1160. [DOI] [PubMed] [Google Scholar]
- [48].Higgs S, Rayner JO, Olson KE, et al. Engineered resistance in aedes aegypti to a West African and a South American strain of yellow fever virus. Am J Trop Med Hyg. 1998;58(5):663–670. [DOI] [PubMed] [Google Scholar]
- [49].Closs O, Aarli JA. Evans blue as counterstain in the demonstration of muscle antibodies by immunofluorescence in myasthenia gravis. J Clin Pathol. 1974;27(2):162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wood E. Molecular cloning. A laboratory manual. Biochem Educ. 1983;11(2):82. [Google Scholar]
- [51].Franz AWE, Sanchez-Vargas I, Adelman ZN, et al. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified aedes aegypti. Proc Natl Acad Sci U S A. 2006;103(11):4198–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Sokal RR, Rohlf FJ. Biometry. The principles and practice of statistics in biological research. 3rd. New York: W.H. Freeman and Co; 1970. Vol. 19. DOI: 10.2307/2412280. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


