ABSTRACT
Eukaryotic cells equip robust systems to respond to stress conditions. In stressed mammalian cells, angiogenin endoribonuclease cleaves anticodon-loops of tRNAs to generate tRNA halves termed tRNA-derived stress-induced RNAs (tiRNAs), which promote stress granule formation and regulate translation. The 5′-tiRNAs (5′-tRNA halves) contain a 2′,3′-cyclic phosphate (cP) and thus belong to cP-containing RNAs (cP-RNAs). The cP-RNAs form a hidden layer of the transcriptome because standard RNA-seq cannot amplify and sequence them. In this study, we performed genome-wide analyses of short cP-RNA transcriptome in oxidative stress-exposed human cells. Using cP-RNA-seq that can specifically sequence cP-RNAs, we identified tiRNAs and numerous other cP-RNAs that are mainly derived from rRNAs and mRNAs. Although tiRNAs were produced from a wide variety of tRNA species, abundant species of tiRNAs were derived from a focal-specific subset of tRNAs. Regarding rRNA- and mRNA-derived cP-RNAs, determination of the processing sites of substrate RNAs revealed highly specific RNA cleavage events between pyrimidines and adenosine in generation of those cP-RNAs. Those cP-RNAs were derived from specific loci of substrate RNAs rather than from the overall region, implying that cP-RNAs are produced by regulated biogenesis pathways and not by random degradation events. We experimentally confirmed the identified sequences to be expressed as cP-RNAs in the cells, and their expressions were upregulated upon induction of oxidative stress. These analyses of the cP-RNA transcriptome unravel an abundant class of short ncRNAs that accumulate in cells under oxidative stress.
KEYWORDS: tRNA; tRNA half; tiRNA; 2′,3′-cyclic phosphate; cP; cP-RNA-seq; oxidative stress
Introduction
tRNAs are universally expressed non-coding RNAs (ncRNAs) that function as fundamental adapter components of translational machinery [1,2]. Besides their role in translation, tRNAs further serve as substrates for shorter ncRNAs. Many organisms produce specific ncRNAs from mature tRNAs or their precursor molecules (pre-tRNAs), and the generated tRNA-derived ncRNAs serve as functional molecules, rather than random degradation products, in various biological processes beyond translation [3–9]. tRNA-derived ncRNAs are classified into two groups: tRNA halves and tRNA-derived fragments (tRFs). tRNA halves range either from the 5′-end to the anticodon-loop (5′-tRNA half) or from the anticodon-loop to the 3′-end (3′-tRNA half) of mature tRNAs. Generally shorter than tRNA halves, tRFs originate from the 5′-part (5′-tRF), 3′-part (3′-tRF), or wholly internal part (i-tRF) of mature tRNAs, or from 5′-leader or 3′-trailer sequences of pre-tRNAs [3–8,10].
Among tRNA-derived ncRNAs, tRNA halves are the most abundant class and their functionality is particularly evident. Eukaryotic tRNA halves were first identified in Tetrahymena as starvation-induced molecules [11]. Their expression was subsequently shown to be triggered by various stress stimuli (e.g., oxidative stress, heat/cold shocks, hypoxia, UV irradiation, low nutrient) in a wide variety of organisms including yeast, plant, and human [12–15], and the stress-induced tRNA halves were termed tRNA-derived stress-induced RNAs (tiRNAs) [16]. In mammalian cells, angiogenin (ANG), a member of the RNase A superfamily, cleaves the anticodon-loops of tRNAs to generate tiRNAs [16,17] (Fig. S1A). In tiRNA biogenesis, stress conditions reduce the levels of RNH1, an ANG-interacting protein that represses ANG activity, which triggers ANG-mediated tRNA cleavage [18]. tiRNA production usually does not affect the levels of mature tRNAs but the generated tiRNAs act as functional molecules. 5′-tiRNAs (5′-tRNA halves) have been shown to regulate translation, promote stress granule formation, enhance immune response, and are implicated in neurodegenerative diseases [15,16,19–24].
While the molecular functions of tiRNAs have been demonstrated, the expression profile of tiRNAs remains elusive. Although production of tiRNAs from a wide variety of tRNAs has been demonstrated by tRNA-specific microarray [18], whole tiRNA sequences have not been well elucidated by next-generation sequencing. Biochemical characterizations have proven that ANG does not hydrolyze a 2′,3′-cyclic phosphate (cP) during its RNA cleavage and thus produces cP-containing RNAs (cP-RNAs) as its final cleavage products [25,26] (Fig. S1A). Indeed, ANG-generated 5′- and 3′-tRNA halves contain a cP and an amino acid at their 3′-end, respectively [27]. Consequently, ANG-generated tRNA halves cannot be ligated to an adapter in standard RNA-seq procedures and therefore they are not efficiently amplified and sequenced by standard RNA-seq [26](Fig. S1B). Standard RNA-seq was used to analyze ANG-induced tRNA fragments [28], but the method is expected to only capture the RNAs containing a phosphate (P) and a hydroxyl group (OH) at their 5′- and 3′-ends, respectively (Fig. S1B). We developed ‘cP-RNA-seq,’ which is able to specifically sequence cP-RNAs [27,29] (Fig. S1B). As 5′-tRNA halves belong to cP-RNAs, we previously applied the method for identification of 5′-tRNA halves expressed in BT-474 human breast cancer cells [27] and BmN4 Bombyx germ cells [30]. Our recent genome-wide analyses of the short cP-RNA transcriptomes using cP-RNA-seq revealed the expression of not only 5′-tRNA halves but also numerous novel cP-RNAs derived from rRNAs and mRNAs expressed in mouse tissues [31]. In this study, we applied cP-RNA-seq to oxidative stress-exposed human cultured cells, revealing a comprehensive picture of tiRNA expression and further elucidating the expression of other short cP-RNA species whose expression is upregulated in stressed human cells.
Results
Sequencing of short cP-RNAs accumulated in oxidative stress-exposed human cell lines
To identify the sequences of tiRNAs and other short cP-RNAs, we used two human cell lines, HeLa and U2OS, whose production of tiRNAs upon induction of oxidative stress, heat shock, and UV irradiation was previously reported [12,13]. For induction of oxidative stress, HeLa and U2OS cells were treated with sodium arsenite (SA) or water (negative control). Generation of tiRNAs was first confirmed by detecting those from cytoplasmic (cyto) tRNAHisGUG, since 5′-tRNAHisGUG half has been shown to be commonly produced as tiRNAs in human, plant, and yeast cells [12]. Although only low levels of 5′-tRNAHisGUG half were observed in control cells, SA treatment enhanced the levels of 5′-tRNAHisGUG half in both HeLa and U2OS cells (Fig. 1A). The levels of mature tRNA were not much affected by the tiRNA production as reported previously [12,16,18].
Figure 1.
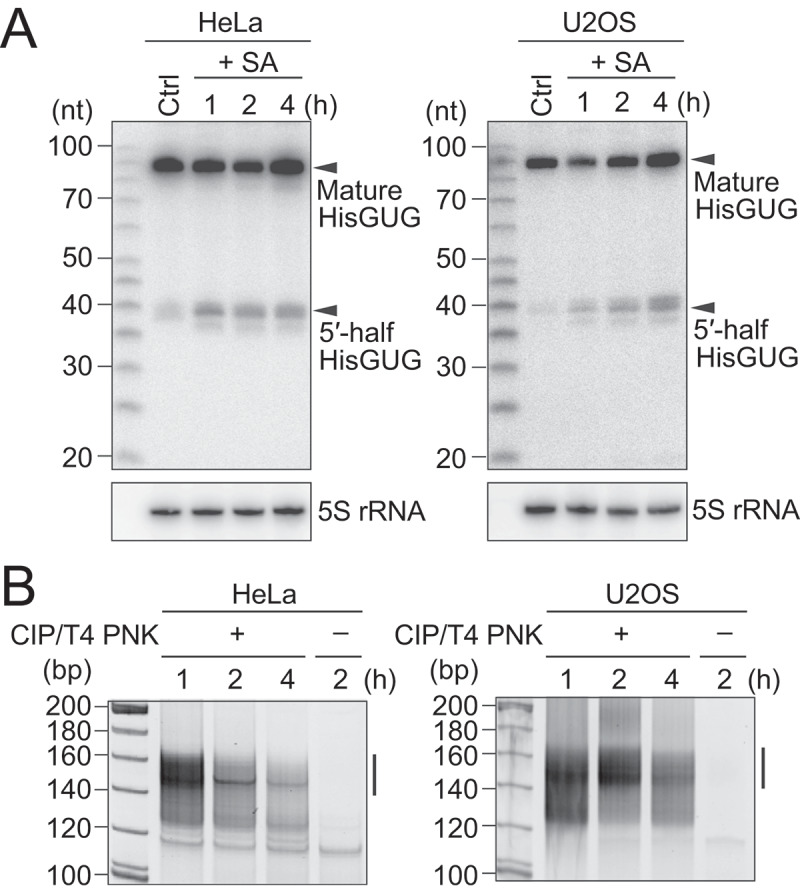
Amplification and sequencing of oxidative stress-induced cP-RNAs.
(A) Total RNAs extracted from SA-treated HeLa or U2OS cells were subjected to northern blots for the 5′-halves of cyto tRNAHisGUG. (B) Gel-purified 20–45-nt RNAs were subjected to cP-RNA-seq, which amplified 140–160-bp cDNA products (5′-adapter, 55 bp; 3′-adapter, 63 bp; and thereby estimated inserted sequences, 22–42 bp).The cDNAs in the region designated with a line were purified and subjected to Illumina sequencing.
To capture an entire picture of 5′-tiRNAs and other short cP-RNAs, 20 to 45-nucleotide (nt) RNAs were gel-purified from SA-treated HeLa and U2OS cells (treated for either 1, 2, or 4 h) and subjected to cP-RNA-seq [27,29]. The method successfully amplified ~140 to 160-bp cDNA products (inserted sequences without adapters were estimated to be 22–42 bp) (Fig. 1B). The cDNA amplifications were dependent on T4 PNK treatment in cP-RNA-seq procedure, indicating that, as expected, the amplified cDNAs were derived from cP-RNAs (Fig. 1B). Illumina sequencing of the amplified cDNAs yielded approximately 45–116 million raw reads, of which >73–83% were extracted as the cP-RNA reads with a length of 20–45 nt (Table S1). Mapping the cP-RNA reads to the ncRNAs and genome revealed that, as in the case of mouse cP-RNAs [31], not only tRNAs but also rRNAs and mRNAs serve as abundant substrates for cP-RNA production (Fig. S2A). Although tRNA-derived reads occupied a substantial proportion of cP-RNAs (4.3–20%), mRNA-derived reads were also abundant (11–16%), and rRNA-derived reads were the most abundant cP-RNA species (31–38%).
tiRNAs are generated from focal cleavage of the anticodon-loop of specific tRNA species
The majority of the identified tRNA-derived ncRNAs were 5′-tRNA halves (5′-tiRNAs) (Fig. 2A). Relative abundance of 5′-tiRNAs in each library was then analyzed, showing consistent overall expression pattern between the six cP-RNA libraries (Fig. 2B). Among 50 human cyto tRNA isoacceptors with different anticodon sequences [32], 5′-tiRNA sequences from 49 isoacceptors were detected in the cP-RNA-seq libraries; the reads of tRNATyrAUA were not detected in any of the six cP-RNA libraries. Although tiRNAs were produced from a wide variety of tRNAs, the majority of the identified 5′-tiRNAs were derived from a rather focused subset of tRNAs, such as cyto tRNALysCUU, tRNAGlyGCC, tRNAGlyCCC, and tRNAGluCUC(Fig. 2B), which were in aggregate the sources of 89–95% of the identified 5′-tiRNAs. The cleavage position of the anticodon-loop for the production of 5′-tiRNAs was predicted by the 3′-terminal nucleotide position of 5′-tiRNAs. As shown in Fig. 2C, the anticodon cleavages majorly occurred between nucleotide position (np; according to the nucleotide numbering system of tRNAs [2]) 34 and 35, and np 36 and 37 in 5′-tiRNA production.
Figure 2.
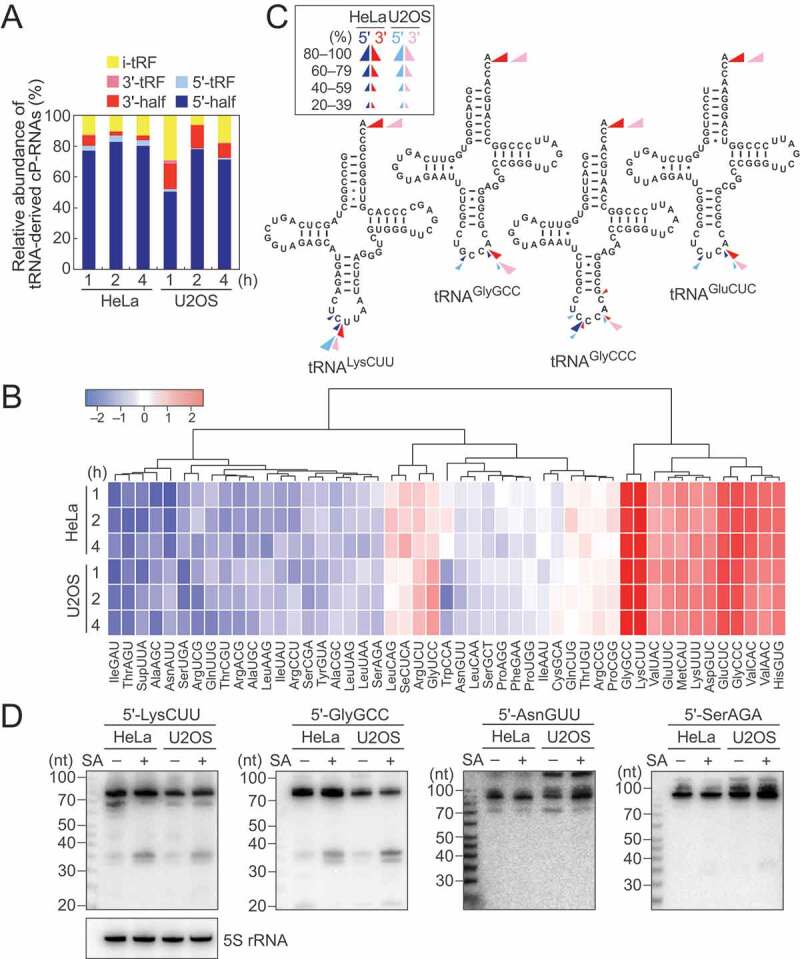
Analyses of tiRNAs.
(A) Proportion of tRNA-derived cP-RNAs classified into the indicated subgroups according to [31]. (B) Expression profiles of the 5′-tiRNA reads derived from respective cyto tRNA isoacceptors. The color key was obtained based on log2 reads per million (RPM) of the 5′-tiRNAs. (C) Cleavage sites in the indicated tRNAs were predicted based on the terminal positions of the identified 5′-tiRNAs (blue arrowheads) and 3′-tiRNAs (red arrowheads). (D) The expression of the indicated 5′-tiRNAs was confirmed by northern blots. The cells treated with SA for 2 h were used.
Among all species of tRNAs, tRNAHisGUG is unique in that it contains an additional nucleotide at np –1 of its 5′-end. A majority of the identified 5′-tiRNAHisGUG contained G–1, but the 5′-tiRNAs containing U–1 or lacking –1 nucleotide (G1) were also substantially detected (Fig. S2B). These heterogeneities are similar to those previously observed in 5′-tRNAHisGUG half molecules in BT-474 human breast cancer cells [33] and mouse tissues [31].
3′-tiRNAs were also detected in our cP-RNA-seq libraries (Fig. 2A). Their 5′- and 3′- terminal nucleotide positions were used for prediction of cleavage sites in 3′-tiRNA biogenesis. As shown in Fig. 2C, the anticodon-loop cleavage sites for 5′- and 3′-tiRNA productions were consistent, suggesting that 5′- and 3′-tiRNAs could be concurrently produced. Indeed, the tRNA species which abundantly generate 5′-tiRNAs, such as cyto tRNALysCUU, tRNAGlyGCC, tRNAGlyCCC, and tRNAGluCUC, were found to be rich sources for 3′-tiRNAs (Fig. S2C). The expression levels of 3′-tiRNAs, especially those with low abundances, did not always correspond to the abundances of 5′-tiRNAs, possibly due to potential amplification/sequencing biases, which were arisen from structures and post-transcriptional modifications of tRNA half molecules, and/or different stabilities of those molecules. As in the case of mouse 3′-tRNA halves detected in cP-RNA-seq [31], 3′-tiRNAs resulted from the cleavage between C75 and A76, as well as anticodon-loop cleavages (Fig. 2C).
To validate the results of sequencing analyses, we selected four 5′-tiRNAs. 5′-tiRNALysCUU and 5′-tiRNAGlyGCC occupied 25–73% and 10–52%, respectively, among all 5′-tiRNA reads and thereby were selected as abundantly identified tiRNAs. On the other hand, 5′-tiRNAAsnGUU and 5′-tiRNASerAGA were selected as tiRNAs with low abundance (occupied 0.0002–0.005% and 0.0002–0.004%, respectively, among all 5′-tiRNA reads). Northern blots showed abundant accumulation of 5′-tiRNALysCUU and 5′-tiRNAGlyGCC in SA-treated cells, while 5′-tiRNAAsnGUU and 5′-tiRNASerAGA showed only faint bands (Fig. 2D). These expression patterns were further observed over the time course of SA treatment (Fig. S3), confirming that our cP-RNA-seq results reflect accumulation of tiRNAs in stress-exposed cells.
To dissect tiRNA expression patterns at isodecoder levels, we focused on the four abundant 5′-tiRNA species: 5′-tiRNALysCUU, 5′-tiRNAGlyGCC, 5′-tiRNAGlyCCC, and 5′-tiRNAGluCUC. Human genome contains 12 isodecoders of cyto tRNALysCUU which are encoded by 17 genes (Fig. S4). Among them, most of the 5′-tiRNAsLysCUU were produced only from three isodecoders: id-1–3 (Fig. 3A). Similarly, other 5′-tiRNAs were also derived from a limited number of specific isodecoders (id-1–4 for 5′-tiRNAGlyGCC; and id-1 and -2 for 5′-tiRNAGlyCCC and 5′-tiRNAGluCUC) (Fig. 3A). The two most abundant isodecoders for each 5′-tiRNA were further subjected to the analyses of their 3′-end positions. In 5′-tiRNALysCUU, relative abundances of the isodecoders ending at np 34 increased over the time course of SA treatment, while the levels of those ending at np 33 decreased (Fig. 3B). This tendency was consistent between id-1 and -2/3, and between HeLa and U2OS cells. Similarly, in other three examined 5′-tiRNAs, specific isodecoders showed changed expression levels over the time course of SA treatment, whose patterns were consistent between different isodecoders and different cell lines (Fig. 3B).
Figure 3.
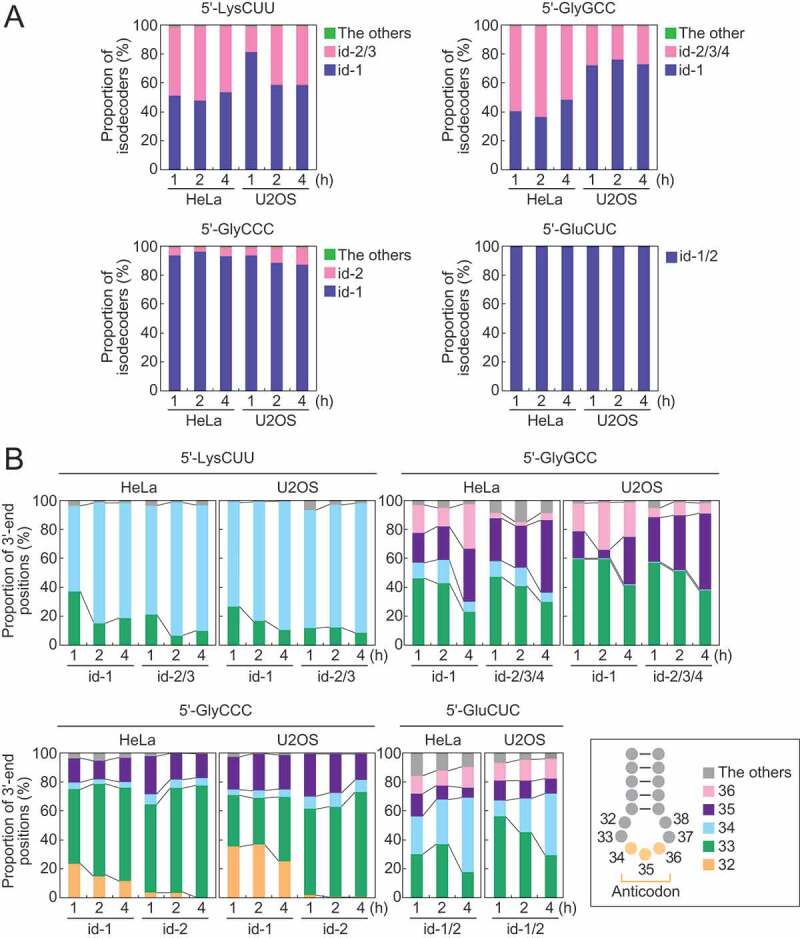
Analyses of tiRNA-producing isodecoders.
(A) Proportion of isodecoders that produce 5′-tiRNA species. (B) The 3′-end positions of the two most abundant isodecoders that produce 5′-tiRNAs.
rRNA-derived cP-RNAs are produced from a specific processing between pyrimidine and adenosine, which are upregulated by oxidative stress
Not only tRNAs but also other RNA species were identified as rich sources of cP-RNAs. rRNA-derived cP-RNAs were the most enriched cP-RNA species in all six cP-RNA libraries (Fig. S2A). Among the four cyto rRNA species (28S, 18S, 5.8S, and 5S), 28S rRNA was the most-rich sources for cP-RNAs (Fig. 4A). Strong nucleotide biases were observed in 5′- and 3′-terminal nucleotides of rRNA-derived cP-RNAs and their adjacent nucleotide on the genome. The 5′-ends were mostly adenosine, whereas the 3′-ends were mostly pyrimidine, and uridine was more enriched compared to cytidine (Fig. 4B). The nucleotides that are one position upstream of the 5′-ends and one position downstream of the 3′-ends also showed clear pyrimidine and adenosine biases (Fig. 4B), strongly suggesting that the rRNA-derived cP-RNAs are produced by an endoribonuclease that specifically recognizes and cleaves rRNAs between cytidine/uridine and adenosine residues.
Figure 4.
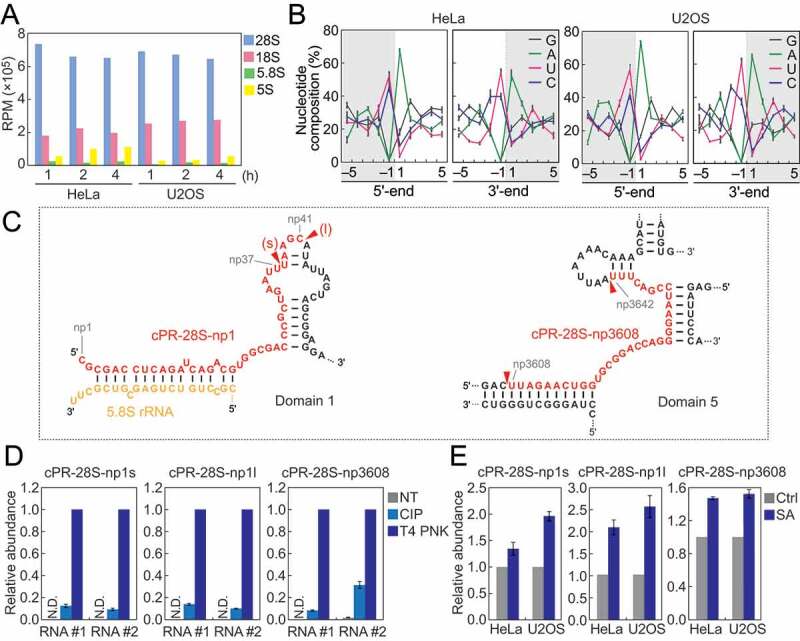
Analyses of rRNA-derived cP-RNAs.
(A) Reads per million (RPM) of cP-RNAs mapped to the indicated rRNAs. (B) Nucleotide compositions around the 5′- and 3′-ends of rRNA-derived cP-RNAs. A dashed line separates upstream and downstream positions for the 5′-and 3′-ends, representing the cleavage site that generates rRNA-derived cP-RNAs (the regions outside of cP-RNA-generating regions are colored in grey). (C) The regions from which the indicated cP-RNAs were derived are shown in red in the secondary structure of substrate rRNAs. The secondary structures are according to [40,41]. (D) The total RNA from SA-treated (2 h) HeLa cells were treated with CIP or T4 PNK and subjected to TaqMan RT-qPCR for detection of the indicated rRNA-derived cP-RNAs. The amounts from T4 PNK-treated RNA were set as 1, and relative amounts are indicated. Averages of three technical replicates with SD values are shown. NT: a non-treated sample used as a negative control. N.D.: not detected. #1/#2: biological replicates. (E) The total RNA from HeLa or U2OS cells treated with SA or water (control) for 2 h were subjected to TaqMan RT-qPCR for detection of the indicated rRNA-derived cP-RNAs. The amounts from control cells were set as 1, and relative amounts are indicated. Averages of three experiments with SD values are shown.
Alignment visualization showed production of specific cP-RNA species from 28S rRNA (Fig. S5), suggesting that the accumulation of cP-RNAs is not just an outcome of degradation but caused by a specific biogenesis mechanism. 5′-terminal region of 28S rRNA was a rich source for cP-RNAs, and short and long forms (termed cPR-28S-np1s and cPR-28S-np1l, respectively) of cP-RNAs were generated from the region (Fig. 4C, S5). A cP-RNA derived from the position 3608–3642 of 28S rRNA was also abundantly detected and thereby termed cPR-28S-np3608 (Fig. 4C, S5). Cleavage positions mostly resided in the loop regions of 28S rRNA, possibly because the regions are highly accessible to endoribonucleases.
To validate that the identified rRNA-derived reads are expressed as cP-RNAs in the cells, HeLa total RNA was treated with calf intestinal phosphatase (CIP; removes 5′-P/3′-P, but not a cP) or T4 polynucleotide kinase (T4 PNK; removes 3′-P/cP), and then we examined the efficiency of the ligations between the three rRNA-derived cP-RNAs (cPR-28S-np1s, cPR-28S-np1l, and cPR-28S-np3608) and a 3′-adapter using TaqMan RT-qPCR as described in [30,31]. As shown in Fig. 4D, all three rRNA-derived cP-RNAs exhibited the highest amplification signals upon T4 PNK treatment, indicating that the major 3′-terminal form of the identified sequences is a cP. CIP treatment showed low levels of signals, suggesting co-expression of 3′-P-containing RNAs as a minor form or occurrence of hydrolysis reaction converting a cP to a P during experimental procedures.
To investigate the relationship between oxidative stress and the expression levels of rRNA-derived cP-RNAs, total RNAs extracted from SA-treated HeLa or U2OS cells were subjected to specific TaqMan RT-qPCR quantification of the three representative rRNA-derived cP-RNAs. As shown in Fig. 4E, in both HeLa and U2OS cells, SA-induced oxidative stress upregulated the levels of all three examined rRNA-derived cP-RNAs, suggesting that the expression of rRNA-derived cP-RNAs is induced by oxidative stress.
mRNA-derived cP-RNAs are produced from a specific processing between uridine and adenosine, which are upregulated by oxidative stress
mRNA-derived cP-RNAs were enriched in cP-RNA libraries at equivalent or even more abundant level to tiRNAs (Fig. S2A). Principal component analysis (PCA) using the reads of mRNA-derived cP-RNAs demonstrated separate clusters of HeLa and U2OS samples (Fig. 5A), suggesting variation of the overall expression profiles of mRNA-derived cP-RNAs in different cells. As in the case of rRNA-derived cP-RNAs, specific nucleotide composition biases were observed in mRNA-derived cP-RNAs. In both HeLa and U2OS cells, the 3′-end and a nucleotide upstream of the 5′-end were strongly biased to pyrimidine, and uridine was particularly enriched compared to cytidine (Fig. 5B). On the other hand, the 5′-end and a nucleotide downstream of the 3′-end were strongly biased to adenosine (Fig. 5B), suggesting that specific cleavages between uridine and adenosine residues produce mRNA-derived cP-RNAs. Alignment visualization showed focal production of specific cP-RNAs from substrate RNAs (Fig. S6), suggesting that the cP-RNA production and accumulation are not caused by random mRNA degradation. Although there are 1,476 ‘UA’ sequences (potential cleavage sites) throughout LPP mRNA (RefSeq ID: NM_005578), most cP-RNAs were produced from one site (position 11,058–11,082; the generated cP-RNA is termed cPR-LPP1) (Fig. 5C, S6). Similarly, SMG1 mRNA (RefSeq ID: NM_015092) contains 1,077 ‘UA’ sequences but a majority of cP-RNAs were derived from one site (position 11,058–11,082; cPR-SMG1 is generated) (Fig. 5C, S6). Our specific TaqMan RT-qPCR succeeded in amplifying and detecting the two-representative mRNA-derived cP-RNAs, cPR-LPP1 and cPR-SMG1, validating their cellular expressions (Fig. 5D). In the detection, T4 PNK treatment of total RNA sample, but not CIP treatment, was required to obtain high amplification signals (Fig. 5D), confirming that the identified RNAs are majorly expressed as cP-RNAs. In both HeLa and U2OS cells, SA-induced oxidative stress upregulated the levels of both mRNA-derived cP-RNAs (Fig. 5E), suggesting that, as in the case of tiRNAs and rRNA-derived cP-RNAs, oxidative stress induces the expression of mRNA-derived cP-RNAs.
Figure 5.
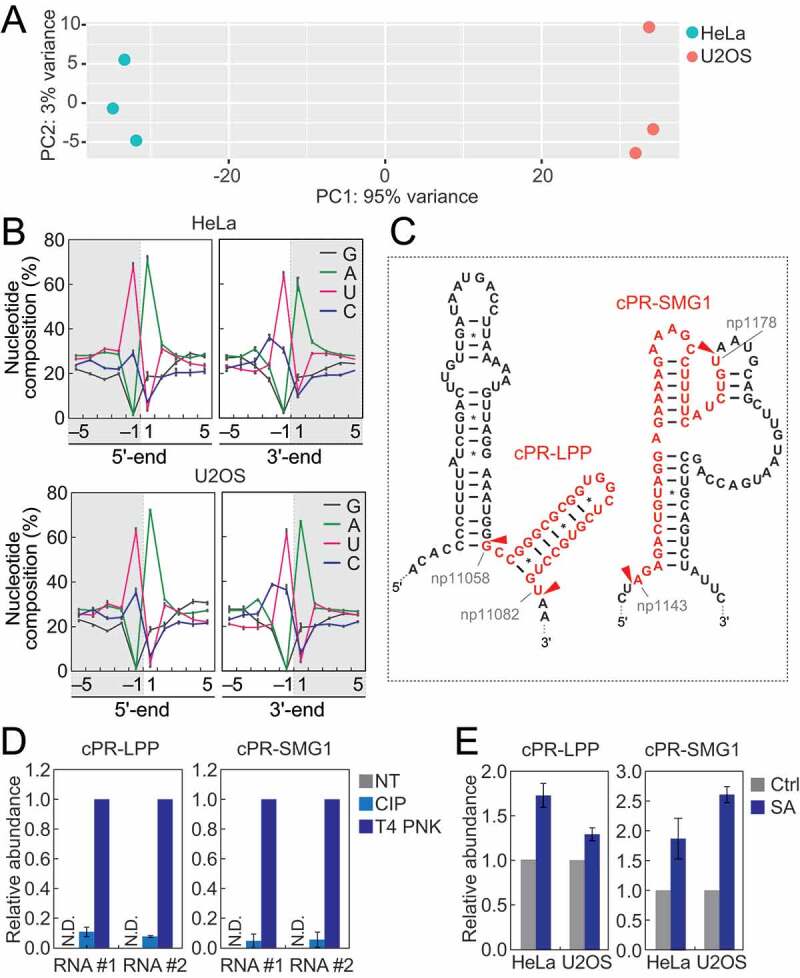
Analyses of mRNA-derived cP-RNAs.
(A) PCA was performed using six individual datasets of mRNA-derived cP-RNAs. RPKM values of substrate mRNAs were obtained and used for the analysis. (B) Nucleotide compositions around the 5′- and 3′-ends of mRNA-derived cP-RNAs. (C) The regions from which the indicated cP-RNAs were derived are shown in red in the secondary structure of the substrate mRNAs. The secondary structures were predicted by ViennaRNA Package 2.0 [42]. (D) The total RNA from the SA-treated (2 h) HeLa cells was treated with CIP or T4 PNK and subjected to TaqMan RT-qPCR for detection of the indicated mRNA-derived cP-RNAs. The amounts from T4 PNK-treated RNA were set as 1, and relative amounts are indicated. Averages of three technical replicates with SD values are shown. NT: a non-treated sample. N.D.: not detected. #1/#2: biological replicates. (E) The total RNA from HeLa or U2OS cells treated with SA or water (control) for 2 h was subjected to TaqMan RT-qPCR for detection of the indicated mRNA-derived cP-RNAs. The amounts from control cells were set as 1, and relative amounts are indicated. Averages of three experiments with SD values are shown.
Discussion
Eukaryotic cells equip robust systems to respond to stress conditions. In this study, we unravelled a previously hidden layer of the transcriptome in oxidative stress-exposed human cells. Because standard RNA-seq cannot sequence cP-RNAs, we utilized cP-RNA-seq to capture primary RNA products of stress-induced ANG cleavage. Our comprehensive capture of short cP-RNA expression profiles revealed the accumulation of tiRNAs and rRNA- and mRNA-derived cP-RNAs in the cells upon induction of oxidative stress. tiRNAs were produced from a wide variety of tRNAs, yet a rather focused subset of tRNAs (e.g., cyto tRNALysCUU, tRNAGlyGCC, tRNAGlyCCC, tRNAGluCUC) became sources for the majority of the identified tiRNAs. Although tRNA modifications that impair reverse transcription can cause biases in sequencing results, the obtained cP-RNA-seq results were consistent with the cellular expression levels of tiRNAs confirmed by northern blot. From the analyses of terminal nucleotide positions of tiRNAs, ANG cleavages at anticodon-loops of tRNAs are predicted to occur between cytidine and uridine/adenosine. Because many other tRNAs also contain 5′-CU-3′ or 5′-CA-3′ sequences at their anticodon-loops, sequence preferences cannot be the sole reason for the abundant production of tiRNAs from specific tRNA species. As tRNA modification states were reported to affect tRNA anticodon cleavage [22,34–36], tRNA structures and/or modifications would affect the amounts of tiRNAs generated from each tRNA. Our analyses on isodecoders suggest that, even from the same substrate tRNAs, tiRNAs with specific lengths are accumulated at different relative abundances over the time course of oxidative stress treatment. Because longer tiRNAs often showed increased abundance, shortenings of tiRNAs by degradation cannot be the reason for the changed relative abundances. The anticodon-loop sites of ANG cleavage might be slightly changed over the time course. Further studies, such as those using in vitro ANG cleavage reaction, are required to fully elucidate the recognition and activity of ANG to tRNAs. The expression levels of mature tRNAs, as well as stability of respective tiRNA species, would also determine the expression levels of tiRNAs. Indeed, tiRNAs are mainly produced from major isodecoders which have high copy numbers in the genome. The tRNA species which were identified as major tiRNA sources have also been identified as abundant sources for mouse 5′-tRNA halves [31], suggesting that the tRNA species particularly susceptible for anticodon cleavage are similar between humans and mice.
In addition to tiRNAs, cP-RNA libraries abundantly contained rRNA- and mRNA-derived sequences that are expected to be generated by specific RNA cleavage between pyrimidine and adenosine. The endoribonuclease generating the rRNA- and mRNA-derived cP-RNAs remains to be determined but should possess highly specific recognition and cleavage activity for the 5′-UA-3′ or 5′-CA-3′ sequences. In human HeLa and U2OS cells, the main cleavages in cP-RNA production occur at 5′-UA-3′, while in mouse tissues, the major cleavage sites are 5′-CA-3′[31]. Because cP-RNAs were not evenly produced from overall sequences of substrate RNAs but rather derived from specific sites, cP-RNA production is not expected to result from random degradation of the substrate RNAs but to be caused by a regulated biogenesis mechanism. The expression of rRNA- and mRNA-derived cP-RNAs was enhanced upon induction of oxidative stress, but the upregulation was less prominent compared to that of tiRNA expression. This could be due to ANG-mediated cleavages of rRNAs and mRNAs being less efficient than those of tRNA anticodons, or rRNA/mRNA-derived cP-RNAs may be generated by other enzyme(s). Considering the already-proven functional significance of tiRNAs, it is not surprising that the identified rRNA- and mRNA-derived cP-RNAs are also expressed as functional molecules. It will be intriguing to examine further the cellular factors that influence, or are influenced by, the expression of rRNA- and mRNA-derived cP-RNAs and their underlying molecular mechanisms, which will define those cP-RNAs as an abundant class of stress-induced functional ncRNAs. tRNA half molecules and rRNA-/mRNA-derived cP-RNAs are more abundantly and constitutively expressed in tissues than in cell lines [17,31], and tRNA half expression can be induced not only by stresses but also by other factors such as sex hormone pathways [27]. It remains to be determined what molecular factors are responsible for the abundant expression of those molecules in tissues and what their physiological roles are.
Material and methods
Cell culture, induction of oxidative stress, and RNA isolation
HeLa and U2OS cells were cultured in DMEM (Life Technologies) containing 10% FBS. The cells were treated with 500 µM of SA (Sigma) to induce oxidative stress. Total RNA from the cells was isolated using TRIsure (Bioline).
Northern blot
Northern blot was performed as previously described [27,37] with the following antisense probes: 5′-tRNAHisGUG half, 5′-GCAGAGTACTAACCACTATACG-3′; 5′-tRNALysCUU half, 5′-GTCTCATGCTCTACCGACTG-3′; 5′-tRNAGlyGCC half, 5′-GAATTCTACCACTGAACCACCAATG-3′; 5′-tRNAAsnGUU half, 5′-GCTAACCGATTGCGCCACA-3′; 5′-tRNASerAGA half, 5′-GTCCATCGCCTTAACCACTC-3′; and 5S rRNA, 5′-GTTCAGGGTGGTATGGCCGT-3′.
cP-RNA-seq and bioinformatics
For cP-RNA-seq, 20–45-nt RNAs were gel-purified and subjected to the cP-RNA-seq procedure as previously described [27,29,30]. The amplified cDNAs were gel-purified and sequenced using the Illumina NextSeq 500 system at the MetaOmics Core Facility of the Sidney Kimmel Cancer Center at Thomas Jefferson University. The sequence libraries contain ~45–116 million raw reads (Table S1) and are publicly available from the NCBI Sequence Read Archive (SRR10309880, SRR10309881, SRR10309882, SRR10309962, SRR10309963, and SRR10309964). Bioinformatic analyses were performed as described previously [31]. In brief, we used the cutadapt tool (DOI: http://dx.doi.org/10.14806/ej.17.1.200) to remove the 3′-adapter sequence. After selecting 20–45-nt reads, we used Rbowtie (1.15.1) [38] for the sequential mappings with one mismatch allowed with reporting one alignment of the best stratum. Reads were mapped to 513 mature cyto tRNAs obtained from GtRNAdb (Release 15) [32], and then to mature rRNAs, to mRNAs, to the mitochondrial genome (NC_012920.1sequence plus 22 mitochondrial tRNA sequences), and to the whole genome (GRCh37/hg19). The expression profiles of 5′-tRNA halves were analyzed by using RPM values (to total reads of 5′-tRNA halves) of each isoacceptor and by using heatmap.2 function of the gplots R package. Our sequencing data can analyse relative abundance of RNAs in each library, and direct quantitative comparison between different libraries could be performed by further experiments such as those using spike-in RNA normalization. Principal component analysis (PCA) of cP-RNA-producing mRNAs was performed by using RPKM values of each mRNA and by using DESeq2 R package.
Quantification of cP-RNAs by TaqMan RT-qPCR
TaqMan RT-qPCR for specific quantification of cP-RNAs was performed as previously described [27]. Briefly, to remove a cP from cP-RNAs, total RNA was treated with T4 PNK, followed by ligation to a 3′-RNA adapter by T4 RNA ligase. Ligated RNA was then subjected to TaqMan RT-qPCR using the One Step PrimeScript RT-PCR Kit (TaKaRa), a TaqMan probe targeting the boundary of the target RNA and 3′-adapter, and specific forward and reverse primers. The quantified cP-RNA levels were normalized to 5S rRNA levels. The sequences of the TaqMan probes and primers are shown in Table S2.
To investigate the terminal structures of the identified cP-RNAs, total RNA was treated with CIP (New England Biolabs) and T4 PNK (New England Biolabs) as previously described [27,39]. The quantified cP-RNA levels were normalized to the 5S rRNA levels that were quantified using the One Step SYBR PrimeScript RT-PCR Kit II (TaKaRa) as described previously [27].
Supplementary Material
Acknowledgments
We are grateful to the members of Kirino lab for helpful discussions. This study was supported in part by the National Institutes of Health Grant (GM106047 and AI130496, to YK), American Cancer Society Research Scholar Grant (RSG-17-059-01-RMC, to YK), and JSPS Postdoctoral Fellowship for Research Abroad (to MS). This study was further supported by the National Institutes of Health Grant (P30CA056036) for utilization of the MetaOmics Core Facility in the Sidney Kimmel Cancer Center at Thomas Jefferson University.
Funding Statement
This work was supported by the American Cancer Society [RSG-17-059-01-RMC]; National Institute of Allergy and Infectious Diseases [AI130496]; National Institute of General Medical Sciences [GM106047].
Disclosure statement
The authors declare that we have no conflicts of interest.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].RajBhandary UL, Soll D.. Transfer RNA in its fourth decade. In: Soll D, RajBhandary UL, editors. tRNA: structure, biosynthesis and function. Washington, D.C: American Society for Microbiology; 1995. p. 1–4. [Google Scholar]
- [2].Sprinzl M, Horn C, Brown M, et al. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sobala A, Hutvagner G. Transfer RNA-derived fragments: origins, processing, and functions. Wiley Interdiscip Rev RNA. 2011;2:853–862. [DOI] [PubMed] [Google Scholar]
- [5].Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013;10:1798–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588:4297–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Shigematsu M, Kirino Y. tRNA-derived short non-coding RNA as interacting partners of argonaute proteins. Gene Regul Syst Bio. 2015;9:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kumar P, Kuscu C, Dutta A. Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem Sci. 2016;41:679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Shigematsu M, Honda S, Kirino Y. Transfer RNA as a source of small functional RNA. J Mol Biol Mol Imag. 2014;1:1–8. [PMC free article] [PubMed] [Google Scholar]
- [10].Pliatsika V, Loher P, Magee R, et al. MINTbase v2.0: a comprehensive database for tRNA-derived fragments that includes nuclear and mitochondrial fragments from all the cancer genome atlas projects. Nucleic Acids Res. 2018;46:D152–D9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lee SR, Collins K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem. 2005;280:42744–42749. [DOI] [PubMed] [Google Scholar]
- [12].Thompson DM, Lu C, Green PJ, et al. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Li Y, Luo J, Zhou H, et al. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008;36:6048–6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hsieh LC, Lin SI, Shih AC, et al. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009;151:2120–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fricker R, Brogli R, Luidalepp H, et al. A tRNA half modulates translation as stress response in Trypanosoma brucei. Nat Commun. 2019;10:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yamasaki S, Ivanov P, Hu GF, et al. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fu H, Feng J, Liu Q, et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–442. [DOI] [PubMed] [Google Scholar]
- [18].Saikia M, Krokowski D, Guan BJ, et al. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J Biol Chem. 2012;287:42708–42725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Emara MM, Ivanov P, Hickman T, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285:10959–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ivanov P, Emara MM, Villen J, et al. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ivanov P, O’Day E, Emara MM, et al. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc Natl Acad Sci U S A. 2014;111:18201–18206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Blanco S, Dietmann S, Flores JV, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. Embo J. 2014;33:2020–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lyons SM, Achorn C, Kedersha NL, et al. YB-1 regulates tiRNA-induced Stress Granule formation but not translational repression. Nucleic Acids Res. 2016;44:6949–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liu S, Chen Y, Ren Y, et al. A tRNA-derived RNA fragment plays an important role in the mechanism of arsenite -induced cellular responses. Sci Rep. 2018;8:16838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Shapiro R, Riordan JF, Vallee BL. Characteristic ribonucleolytic activity of human angiogenin. Biochemistry. 1986;25:3527–3532. [DOI] [PubMed] [Google Scholar]
- [26].Shigematsu M, Kawamura T, Kirino Y. Generation of 2ʹ,3ʹ-Cyclic Phosphate-Containing RNAs as a Hidden Layer of the Transcriptome. Front Genet. 2018;9:562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Honda S, Loher P, Shigematsu M, et al. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci U S A. 2015;112:E3816–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Su Z, Kuscu C, Malik A, et al. Angiogenin generates specific stress-induced tRNA halves and is not involved in tRF-3-mediated gene silencing. J Biol Chem. 2019;294:16930–16941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Honda S, Morichika K, Kirino Y. Selective amplification and sequencing of cyclic phosphate-containing RNAs by the cP-RNA-seq method. Nat Protoc. 2016;11:476–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Honda S, Kawamura T, Loher P, et al. The biogenesis pathway of tRNA-derived piRNAs in Bombyx germ cells. Nucleic Acids Res. 2017;45:9108–9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Shigematsu M, Morichika K, Kawamura T, et al. Genome-wide identification of short 2ʹ,3ʹ-cyclic phosphate-containing RNAs and their regulation in aging. PLoS Genet. 2019;15:e1008469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shigematsu M, Kirino Y. 5ʹ-Terminal nucleotide variations in human cytoplasmic tRNAHisGUG and its 5ʹ-halves. RNA. 2017;23:161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang X, Matuszek Z, Huang Y, et al. Queuosine modification protects cognate tRNAs against ribonuclease cleavage. RNA. 2018;24:1305–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Schaefer M, Pollex T, Hanna K, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lyons SM, Fay MM, Ivanov P. The role of RNA modifications in the regulation of tRNA cleavage. FEBS Lett. 2018;592:2828–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Honda S, Kirino Y, Maragkakis M, et al. Mitochondrial protein BmPAPI modulates the length of mature piRNAs. Rna. 2013;19:1405–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Honda S, Loher P, Morichika K, et al. Increasing cell density globally enhances the biogenesis of Piwi-interacting RNAs in Bombyx mori germ cells. Sci Rep. 2017;7:4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gorski JL, Gonzalez IL, Schmickel RD. The secondary structure of human 28S rRNA: the structure and evolution of a mosaic rRNA gene. J Mol Evol. 1987;24:236–251. [DOI] [PubMed] [Google Scholar]
- [41].Petrov AS, Bernier CR, Gulen B, et al. Secondary structures of rRNAs from all three domains of life. PLoS One. 2014;9:e88222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lorenz R, Bernhart SH, Honer Zu Siederdissen C, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


