Fig. 2.
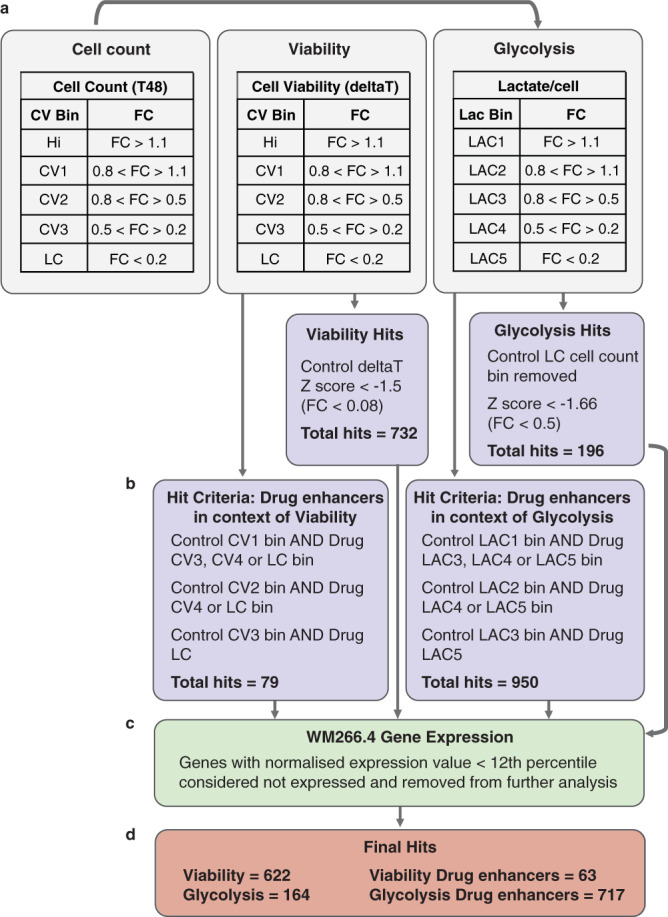
Hit classification strategy applied to the genome wide primary screen. Genes were classified according to a binning strategy for each of the screen output parameters to identify regulators of cell viability and glycolysis (a). Control and drug hit bins were then applied to identify genes that enhance drug effects on viability and glycolysis. Genes that satisfied the listed control and drug bin criteria were classified as drug enhancer hits. (b). Putative hits were triaged for expression in the WM266.4 cell line (c), resulting in 622 viability hits, 164 glycolysis hits, 63 viability drug enhancer hits and 717 glycolysis drug enhancer hits (d) (FC = fold change; Hi = high count; CV = cell viability; LC = low count; LAC = lactate).
