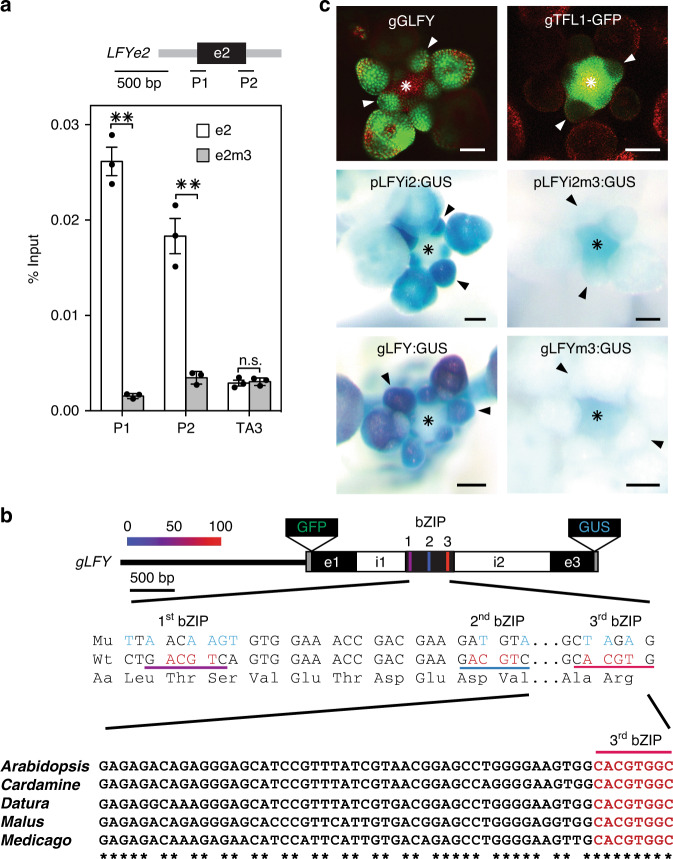
Fig. 2. Exonic bZIP cis motifs mediate TFL1 recruitment to LFY.
a TFL1 recruitment to LFY exon 2 (e2) or a bZIP-binding site mutated version thereof (e2m3) in 42-day-old short-day-grown plants. Top: LFY exon 2 (e2, black rectangle) and T-DNA vector (grey line). Centre: location of amplicons P1 and P2, each consisting of one exon 2-specific and one vector-specific primer. Bottom: gTFL1-GFP ChIP-qPCR. Progeny pools of >50 random gTFL1-GFP T1 plants transformed with e2 or e2m3 were analyzed. Shown are mean ± SEM of three independent biological experiments (black dots). P values unpaired one-tailed t-test: **P1 = 0.0018, **P2 = 0.0078; n.s. TA3 = 0.35. b Top: genomic LFY construct with GFP (gGLFY) or beta-glucuronidase (gLFY-GUS). Putative bZIP-binding motifs in LFY exon 2 are colour-coded based on conservation. For the pLFYi2:GUS reporter diagram see Supplementary Fig. 6a. Centre: mutation of the three bZIP-binding motifs without changing the primary amino acid sequence. Bottom: evolutionary conservation of the G-box (3rd bZIP). c Top: expression domain of LFY and TFL1 proteins in inflorescence apices with flower primordia. Centre and Bottom: accumulation of beta-glucuronidase in a wild-type or bZIP-binding site mutated (m3) reporter (pLFYi2:GUS) or genomic construct (gLFY-GUS). Plants were grown in long day. Staining was conducted under identical conditions. Arrowheads: flower primordia; asterisk: inflorescence shoot apex; Scale bars: 2 mm. See also Supplementary Figs. 4–6.