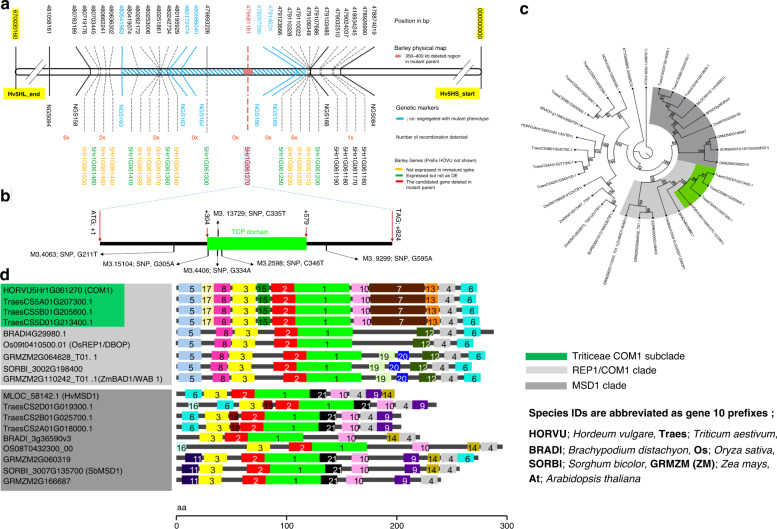
Fig. 2. Map-based cloning, phylogenetic analysis, and protein structural variation of COM1.
a Physical and genetic maps of com1.a from 100 recombinant plants or ~6,000 gametes. A single gene (red; HORVU5Hr1G061270, a single-exon TCP transcription factor) was the strongest candidate and deleted in the mutant parent com1.a. b COM1 gene model containing one TCP DNA binding domain (green box). Six barley TILLING alleles are shown with prefix M3. c UPGMA phylogenetic tree, using 1,000 bootstrap replications, of COM1 homologs (highlighted in light gray) and paralogs (in dark gray) appeared as first- and second-best hits, respectively, in the blast search. Bootstrap values (in percentage) are shown within the circular cladogram along the edges of the branches. d Evolutionarily conserved motifs, among COM1 homologous and paralogous proteins (presented as phylogenetic tree in c) using the tool SALAD. Each colored box represents a different and numbered protein motif. For example, motif 1 in light green represents the TCP domain. Motifs 7, 13, 15, and 17 of the REP1/COM1 clade are specific to the Triticeae. (see also Supplementary Fig. 6). .