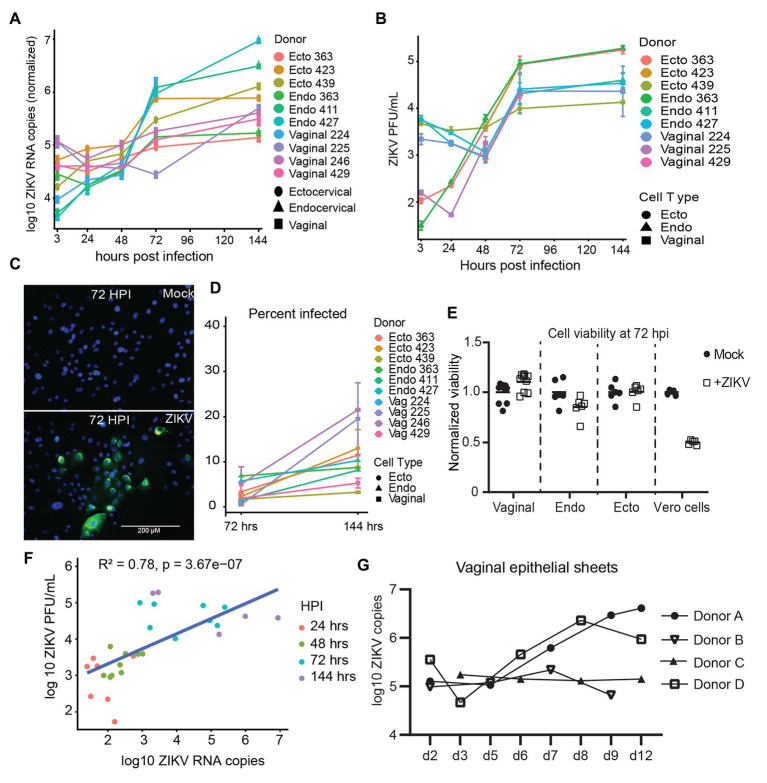
Figure 1.
Kinetics of Zika virus (ZIKV) replication in female genital tract epithelial cells and tissue. Epithelial cells were exposed to ZIKV at a multiplicity of infection (MOI) of 1 for 1.5 h, washed, and then cultured. At the indicated time points, cells were processed for quantification of ZIKV infection by multiple methods. (A) RNA from infected cells was reversed transcribed, and copies of ZIKV virus genome and cellular housekeeper RPP30 were quantified by droplet digital PCR (ddPCR). Y-axis indicates the log10 copy number of ZIKV genomes detected in 200 ng of input RNA, normalized to RPP30 levels. Each line is the average results for duplicate wells, error bars are standard deviation, and each color is a separate experiment. (B) Supernatants from infected cells were added to Vero cells for plaque assays as described in Materials and Methods section. Plaque-forming units (PFU) per ml of supernatant averaged over three separate wells are plotted; error bars are standard deviation. (C) Staining for ZIKV envelope protein. At 72 hpi, cells were fixed in 4% paraformaldehyde, permeabilized, stained with anti-flavivirus envelope 4G2, and counterstained with DAPI. One representative of nine separate experiments is shown, (D) Kinetics of the percent of ZIKV-infected cells. At least 500 DAPI+ cells per well were analyzed for ZIKV envelope E protein expression to calculate the percentage of infected cells. Each point is the average of two separate wells, error bars are standard deviation, and each line is a separate experiment. (E) Cell viability was assessed with the Viral ToxGlo assay. Fold difference from the mean of values for mock infected cells is plotted. Vero cells were infected with ZIKV at an MOI of 0.05 and assessed for cytotoxicity at 72 hpi as a positive control. Each point is a separate well. (F) Correlation between ZIKV RNA copies measured by ddPCR and ZIKV PFUs as determined by plaque assay. Data generated from at 48, 72, and 144 hpi time points were pooled for the regression. (G) Epithelial sheets from vaginal tissue were exposed to 500,000 PFU of ZIKV overnight, extensively washed, and cultured for the indicated times. At harvest, total tissue RNA was extracted, and ZIKV genomes quantified by ddPCR and normalized to RPP30 levels. Points are the average of two or three independent wells per experiment/donor.