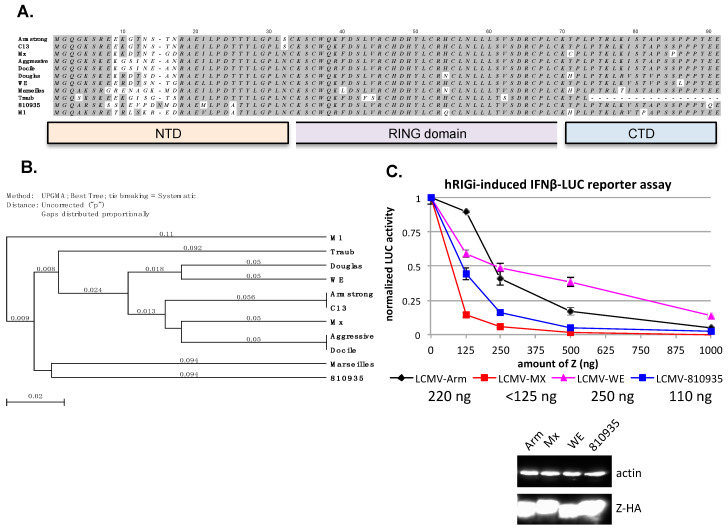
Figure 4.
Comparative hRIG-I inhibitory analysis of Z proteins from different LCMV strains. (A) The amino acid sequences of Z proteins from different LCMV strains, Armstrong (P18541.3), cl13 (ABC96003), Mx (CAA10342), Aggressive (ACA61299), Docile (ACA61303), Douglas (ACV72588), WE, Marseille #12 (ABB88930), Traub (P19325, partial sequence), 810935(ACV72580), and M1 (BAJ52729) were aligned using the MacVector software, with the N-terminal domain (NTD), the central RING domain, and the C-terminal domain (CTD) shown at the bottom. (B) The phylogenetic tree based on the amino acid sequences of Z proteins. (C) Comparative hRIG-I inhibitory effects by the respective Z proteins (in a dose-dependent manner) of four different LCMV strains (Armstrong, WE, Mx, and 810935] were determined as described in the legend of Figure 2. Notable differences in the size of the Z proteins correspond to differences in their primary amino acid sequences. The IC50 values for each of the LCMV Z proteins tested are shown in the figure legend. Western blot analysis of LCMV Z proteins is shown.