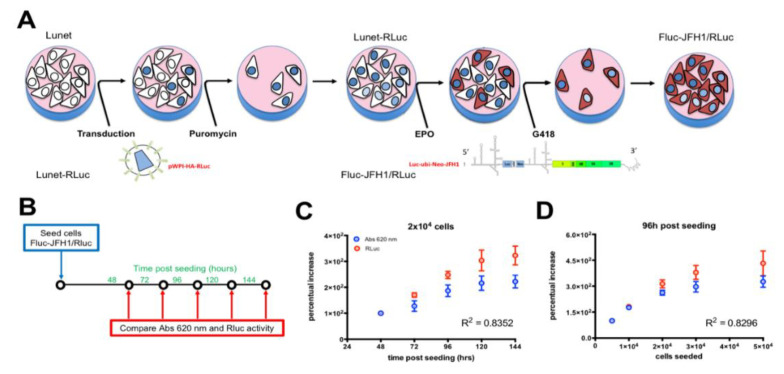
Figure 2.
Generation of an HCV subgenomic double reporter replicon cell line allows accurate quantification of cell number. (A) Lunet cells were transduced with pWPI-HA-RLuc, mediating expression of the HA-RLuc reporter under control of the EF1α promoter. Transduced cells were selected using Puromycin to generate Lunet-RLuc cells, allowing to easily monitor cell number and viability by firefly luciferase assays. Subsequently cells were electroporated with in vitro transcribed RNA from plasmid pFKI389Luc-ubi-neo/NS3-3′_dg_JFH. Cells stably replicating HCV subgenomic RNA were selected using G418 (750 µg/mL) to generate FLuc-JFH1/RLuc cells, allowing to simultaneously monitor cell number and viral replication by dual luciferase assays. (B). Different amounts of FLuc-JFH1/RLuc cells were seeded either in 96- (for MTT assays) or 24- (for RLuc assays) well plates and incubated for the indicated time points before being processed as described in the Materials and Methods section. (C) Measurements relative to 2 × 104 cells seeded/cm2, cultured for the indicated time, are expressed as the percentage of the signals obtained 48 h post-seeding. The R2 relative to the linear regression between Abs at 620 nm (blue circles) and RLuc values (red circles) is shown. Data are mean + standard error of the mean relative to three independent experiments. (D) Data relative to the indicated number of cells seeded/cm2 and cultured for 96 h, are expressed as the percentage of the signals obtained for 5 × 103 cells seeded/cm2. The R2 relative to the linear regression between Abs 620 nm (blue circles) and RLuc values (red circles) is shown. Data are mean ± standard error of the mean relative to three independent experiments.