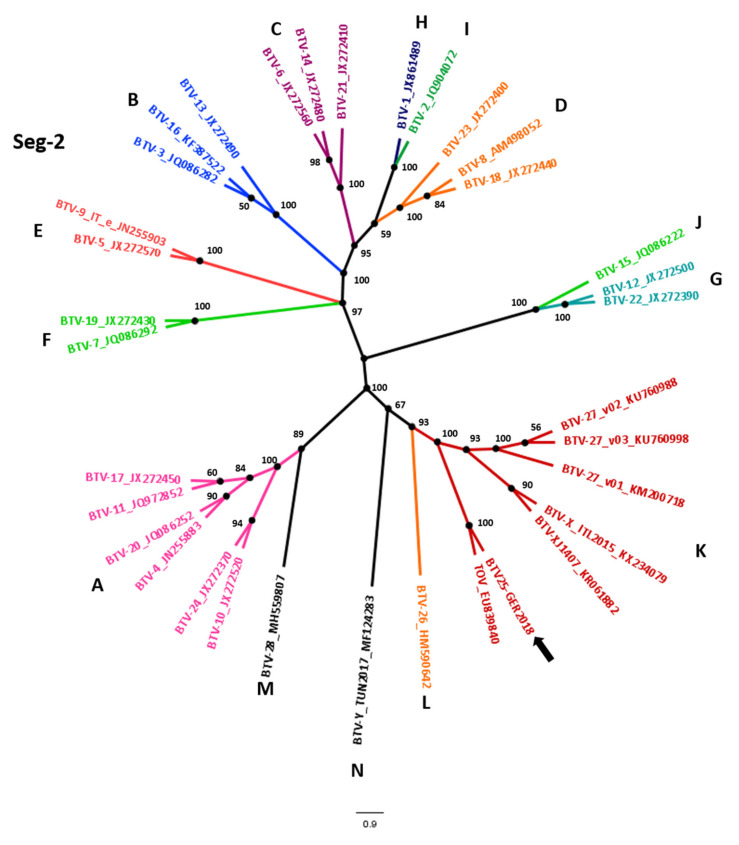
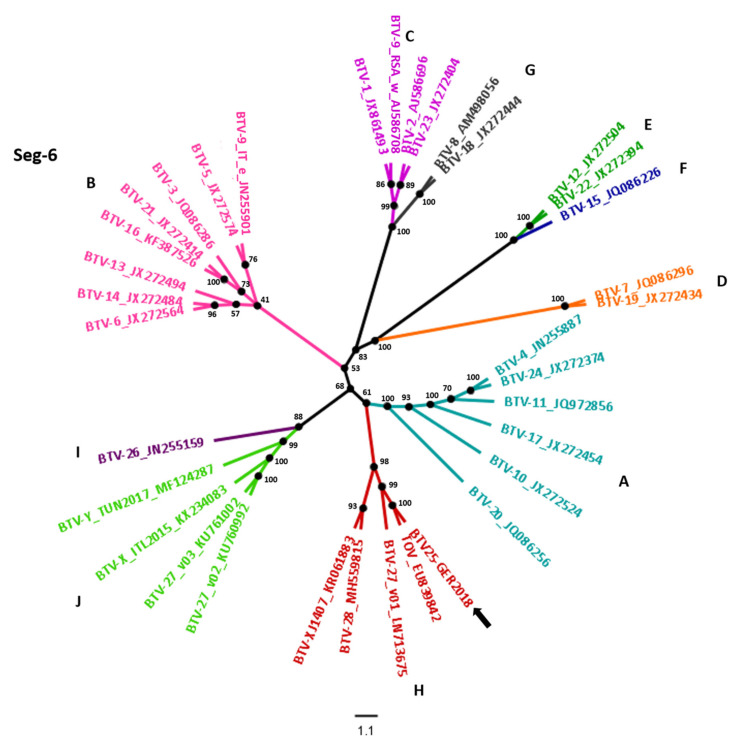
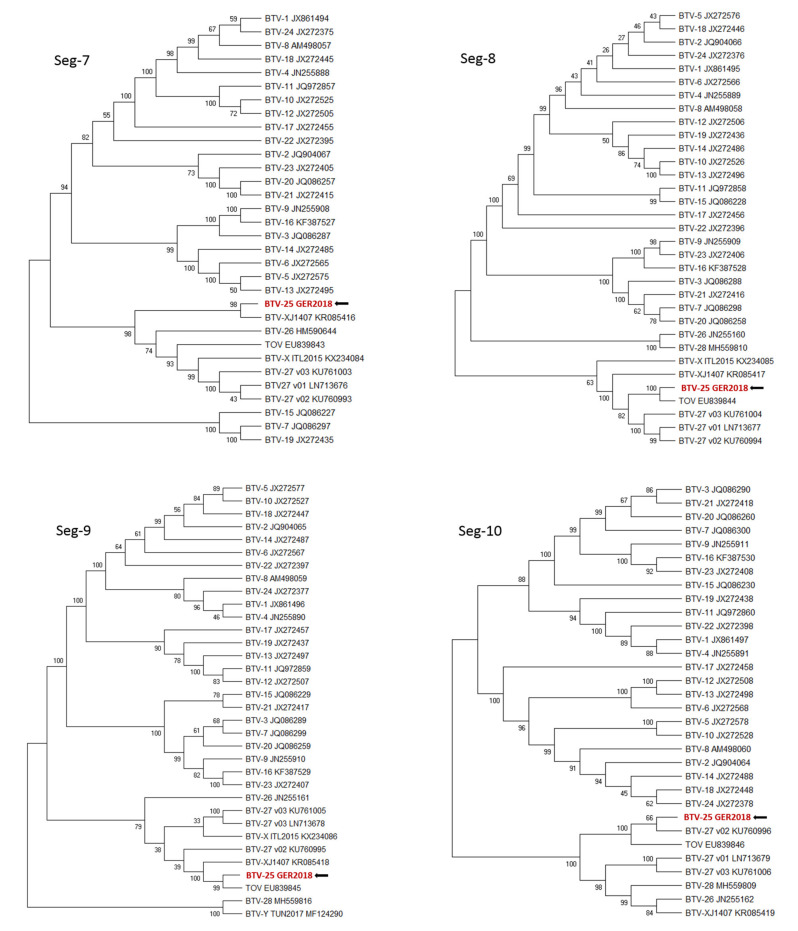
Figure 1.
Phylogenetic analyses of the BTV-25-GER2018 genome. The phylogenetic trees of each of the 10 segments were created with MegaX using the genetic distinction model Tamura–Nei and tree-built method UPGMA including BTV strains representing the known BTV serotypes published [6]. We performed a bootstrap analysis with 1000 replications. The colors of the phylogenetic trees of segment 2 (Seg-2) and segment 6 (Seg-6) represent the different nucleotype groups [32] and trees were modified with FigTrees. For easier identification of the different nucleotypes A-N, the unrooted tree layout was chosen for Seg-2 and Seg-6. The arrows point at the BTV-25-GER2018 sequence.