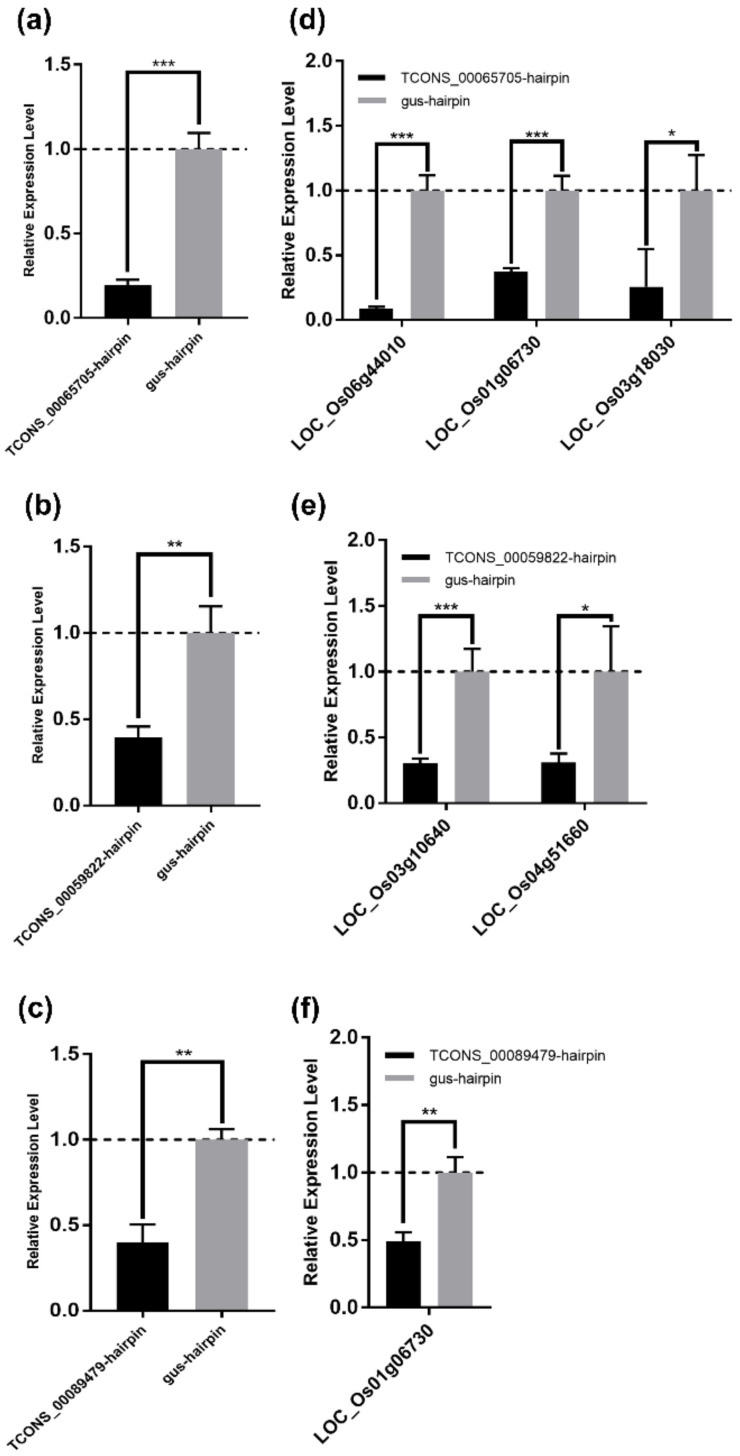
Figure 8.
Experimental validation of the lncRNA–mRNA regulatory relationship in the DElncRNA–DEmRNA co-expression network. (a–c) The silencing of the lncRNA TCONS_00065705 (a), TCONS_00059822 (b), and TCONS_00089479 (c) was confirmed by qRT-PCR. Total RNA of rice callus was extracted at 7 days after Agrobacterium infection. OsUBC gene served as an internal reference for the relative quantification. The values represent means of the lncRNA expression levels ± standard deviations (SD) relative to the gus-hairpin-expressed callus tissues (n = 3 biological replicates). Data were analyzed by the Student’s t-test, and asterisks denote significant differences between lncRNA-silenced and gus-hairpin-expressed callus tissues (two-sided, ** p < 0.01, *** p < 0.001). (d–f) qRT-PCR analyses of the mRNA co-expressed with TCONS_00065705 (d); TCONS_00059822 (e); and TCONS_00089479 (f). Total RNA in (a–c) was used for qRT-PCR analyses. OsUBC gene served as an internal reference for the relative quantification. The values represent means of the mRNA expression levels ± standard deviations (SD) relative to the gus-hairpin-expressed callus tissues (n = 3 biological replicates). Data were analyzed by the Student’s t-test, and asterisks denote significant differences between lncRNA-silenced and gus-hairpin-expressed callus tissues. (two-sided, * p < 0.05, ** p < 0.01, *** p < 0.001).