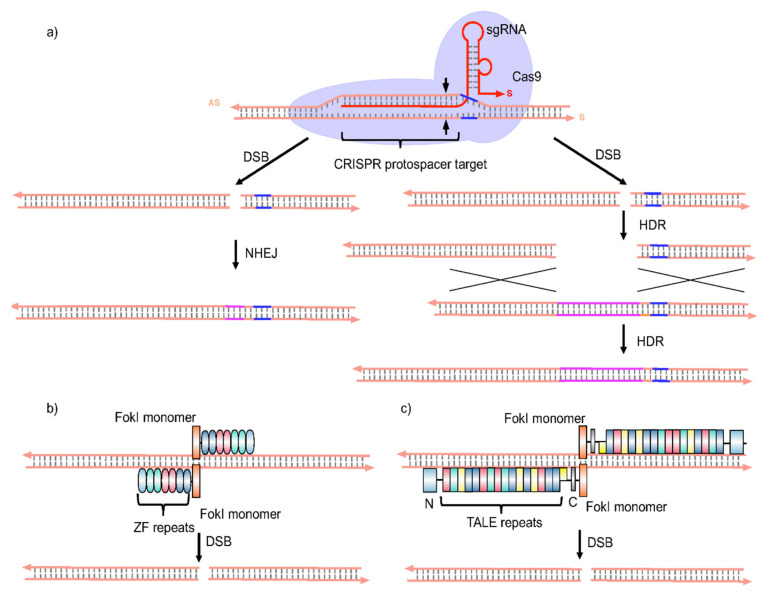
Figure 6.
Schematic representations of gene editing technologies. (a) Class 2 clustered regularly interspaced short palindromic repeat (CRISPR) systems are part of an adaptive immune system in bacteria Due to comparative simplicity and adaptability, CRISPR has rapidly become a popular genome engineering approach. CRISPR-associated endonuclease (Cas protein, typically Cas9 from S. pyogenes) is a nuclease capable of creating targeted double-strand breaks (DSBs) when directed to a given DNA locus by means of a guide RNA (gRNA or sgRNA, red strand in figure). The sgRNA is a short stretch of synthetic RNA composed of a scaffold sequence at its 3′-end for binding to Cas9 and a user-defined sequence (20–22 nts) at its 5′-end, for binding to the AS strand of a selected CRISPR protospacer target in DNA. This CRISPR protospacer target should be immediately adjacent to a protospacer adjacent motif (PAM) (typically 5′-NGG in the DNA S strand, blue strand region in figure). The CRISPR/Cas9 system operates when Cas9 and the sgRNA form a ribonucleoprotein complex. The sgRNA scaffold sequence interacts with surface-exposed positively-charged grooves on Cas9, and the resulting complex then undergoes a conformational change that enables the sgRNA protospacer to zip-bind with the AS strand of the CRISPR protospacer target sequence in a 3′ to 5′ direction, starting with the gRNA seed sequence (first 8–10 nts). Assuming seed sequence/AS strand complementarity, the sgRNA will then continue to anneal to the end, thereby enabling the Cas9 functional endonuclease domains (RuvC and HNH) to undergo a second conformational change that leads to a double-strand break (DSB) (~3–4 nts upstream of the PAM sequence). The resulting, highly selective DSB is then repaired by one of two general repair pathways; (1) The efficient but error-prone non-homologous end joining (NHEJ), (2) The less efficient but high-fidelity homology-directed repair (HDR). The NHEJ repair pathway causes small nucleotide insertions or deletions (indels) at the DSB site. In most cases, indels result in frameshift mutations leading to premature stop codons within the targeted gene and may result in amino acid deletions, insertions and/or protein loss-of-function at the level of translation. Accordingly, the NHEJ repair pathway can be used to disrupt the open reading frame of a gene and generate a knock-out (KO) allele to a target gene that bears the selected protospacer target sequence. On the other hand, the HDR pathway can be used to integrate a donor DNA sequence into the DSB site to create a precise deletion, substitution, or insertion, that leads either to the correction of a pathologic gene or else the targeted knock-in (KI) of a DNA fragment or new gene of interest. Cys2His2 zinc fingers (ZFs) are DNA-binding domains that each recognize approx. three bps of DNA. (b) Alteration of a small number of amino acid residues in or near an α-helix within this domain can lead to changes in DNA-binding specificity. Engineered zinc fingers can be joined together into more extended arrays capable of recognizing longer DNA sequences. A large number of zinc finger arrays engineered can be fused to a non-specific nuclease domain from the Type IIS FokI restriction enzyme to create zinc finger nucleases (ZFNs). The FokI nuclease functions as a dimer, and therefore two zinc finger arrays must be designed for each target site. Most recent ZFN pairs contain complementary obligate heterodimeric FokI domains. A ZFN pair is shown to produce a highly selective DSB, that is followed by NHEJ or HDR as appropriate. Transcription activator-like effector nucleases (TALENs) have rapidly emerged as an alternative to ZFNs. (c) TALENs are similar to ZFNs and comprise a non-specific FokI nuclease domain fused to a customizable DNA-binding domain. The fundamental building blocks used to create the DNA-binding domain of TALENs are highly conserved repeats derived from naturally occurring transcription activator-like effectors (TALEs) encoded for by Xanthomonas proteobacteria. DNA binding by TALEs is mediated by arrays of highly conserved 33–35 amino acid residue repeats flanked by additional TALE-derived domains at the amino- and carboxy-terminal ends of a given array. Individual repeats bind a single nucleotide residue of DNA as determined by the identities of two hypervariable residues typically found at amino acid residue positions 12 and 13 in each TALE repeat. TALE repeats with hypervariable residues N & N (green) recognize G nucleotide residues, N & I (yellow) recognize A nucleotide residues, H & D (purple) C nucleotide residues, and N & G (red) T nucleotide residues, respectively. As in b), the FokI nuclease functions as a dimer, and therefore two TALENs must be designed for each target site. A TALEN pair may then produce a highly selective DSB, that is followed by NHEJ or HDR as appropriate.