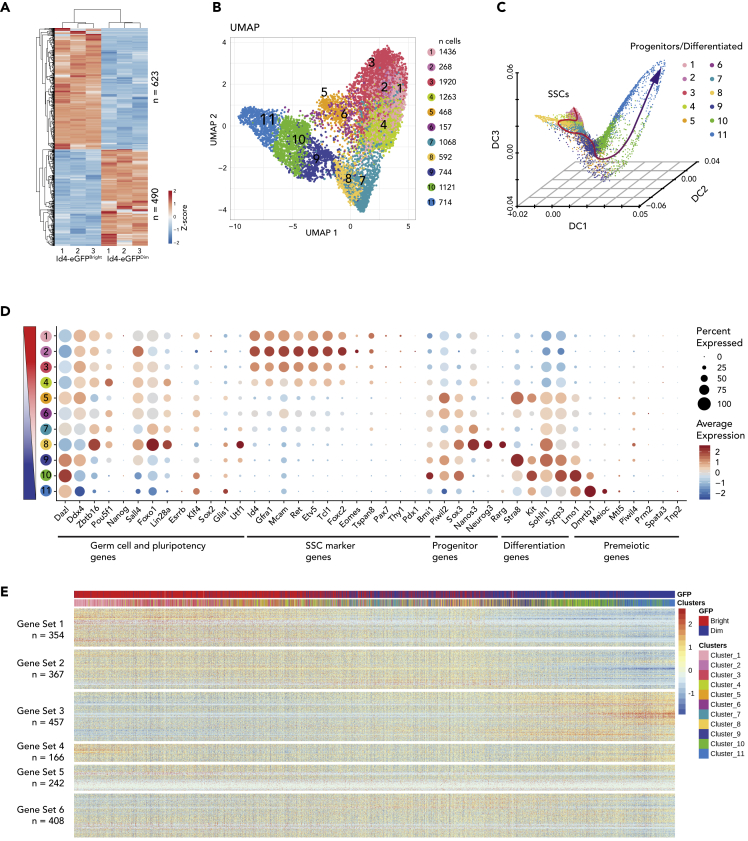
Figure 1.
Differential Gene Expression in SSC-Enriched and Progenitor-Enriched Spermatogonial Subpopulations
(A) Differential gene expression profiling of ID4-eGFPBright and ID4-eGFPDim spermatogonia by bulk RNA-seq. Genes with >1.5 LFC (Log2 Fold Change difference >1.5, p < 0.01) were hierarchically clustered. Red–blue colors indicate high–low expression levels in Z score. Gene-specific results of bulk RNA-seq can be found in Table S1.
(B) Subpopulations among P6 ID4-eGFPBright and ID4-eGFPDim spermatogonia are revealed in this UMAP display of single-cell RNA-seq (scRNA-seq) data. Each dot represents a single cell. The cell number in each cluster is indicated via color coding.
(C) Developmental trajectory of neonatal spermatogonia. A dynamic diffusion map shows the developmental progression of ID4-eGFPBright to ID4-eGFPDim spermatogonia in pseudotime.
(D) Scaled average expression of selected marker genes and the percentage of cells within each cluster with detectable expression (dot radius) for each selected marker gene within each cell cluster. The cell cluster order was arranged by Diffusion Map from top to bottom.
(E) Scaled, normalized expression of the 2,000 most variably expressed genes in ID4-eGFPBright and ID4-eGFPDim spermatogonia. Cells are ordered along their pseudotime developmental progression shown in Figure 1C. The top bar indicates the progression from ID4-eGFPBright (red) to ID4-eGFPDim (blue) cells based on intensity of ID4-eGFP expression. The lower bar represents the cell clusters identified according to the color code shown in Figure 1B. These 2,000 most variably expressed genes were hierarchically clustered into six gene sets based on expression patterns throughout the pseudotime progression. A detailed list of variably expressed genes is provided in Table S1.