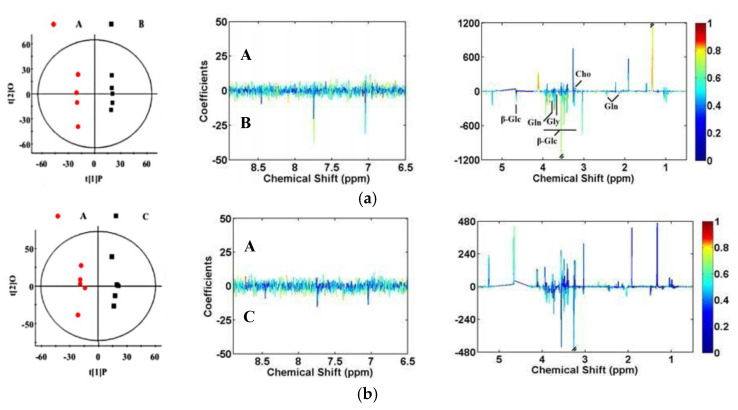
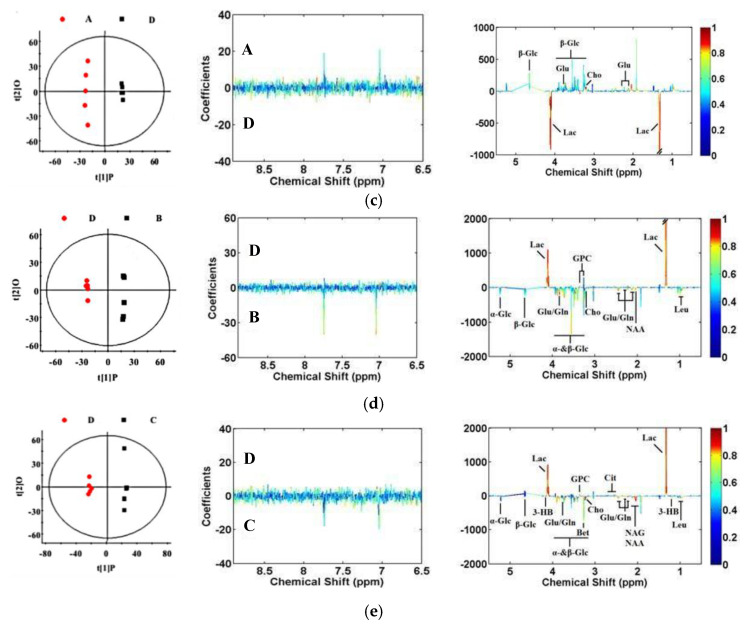
Figure 3.
Orthogonal projection to latent structure with discriminant analysis (OPLS-DA) scores plots (left panel) derived from 1H NMR spectra of serum and corresponding coefficient loading plots (right panel) were obtained from the (A) CON group, (B) HLB group, (C) RSM group and (D) HLB + RSM group. Score plot of serum: (a): CON group vs. HLB group, R2X = 22.5%, and Q2 = −0.077; (b): CON group vs. RSM group, R2X = 25.8%, and Q2 = −0.480; (c): CON group vs. HLB + RSM group, R2X = 26.2%, and Q2 = −0.241; (d): HLB + RSM group vs. HLB group, R2X = 24.7%, and Q2 = −0.503; (e): HLB + RSM group vs. RSM group, R2X = 28.4%, and Q2 = −0.341. Keys were the same as shown in Figure 1. The color map shows the significance of metabolites variations between the two classes. Peaks in the positive direction indicate that the metabolites are more abundant in the groups in the positive direction of the first principal component. Therefore, peaks in the negative direction indicate that metabolites that are more abundant in the groups in the negative direction of the first principal component.