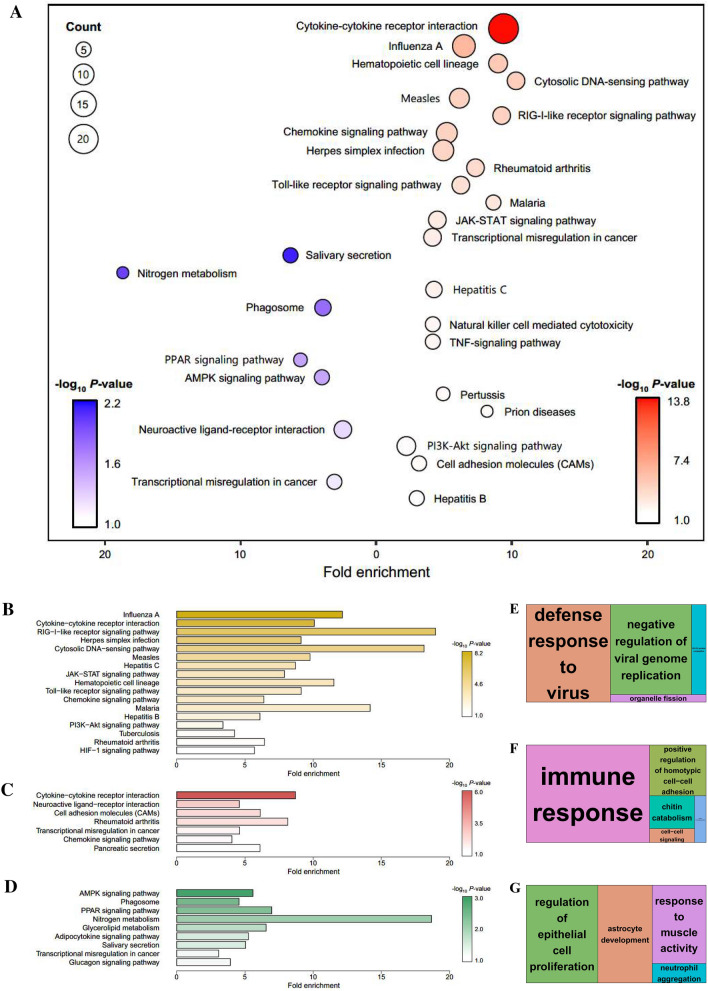
Figure 4.
Enrichment analyses based on the DAVID database, aimed at discovering the biological meaning of representative up- and down-regulated signalling pathways following PRRSV infection. A–G Whole GCN network (A), 3 dpi-specific network (cluster 1; B and E), lung-specific network (clusters 2 and 3; C and F), and BLN-specific network (clusters 4 and 5; D and G) were used for the analyses. KEGG-enriched pathways for the whole GCN network (A) and each specific network (B–D) were visualised by generating bubble and bar plots. A bubble plot corresponding to the whole network was generated for each analysis for the up-regulated (red) and down-regulated genes (blue) (A).
Data information: Significantly enriched pathways represented in the plots met the following cut-off criterion: − log10 P value > 1.0. GO treemaps were created based on the P values associated with the BP terms for each specific network (E–G).