Figure 3.
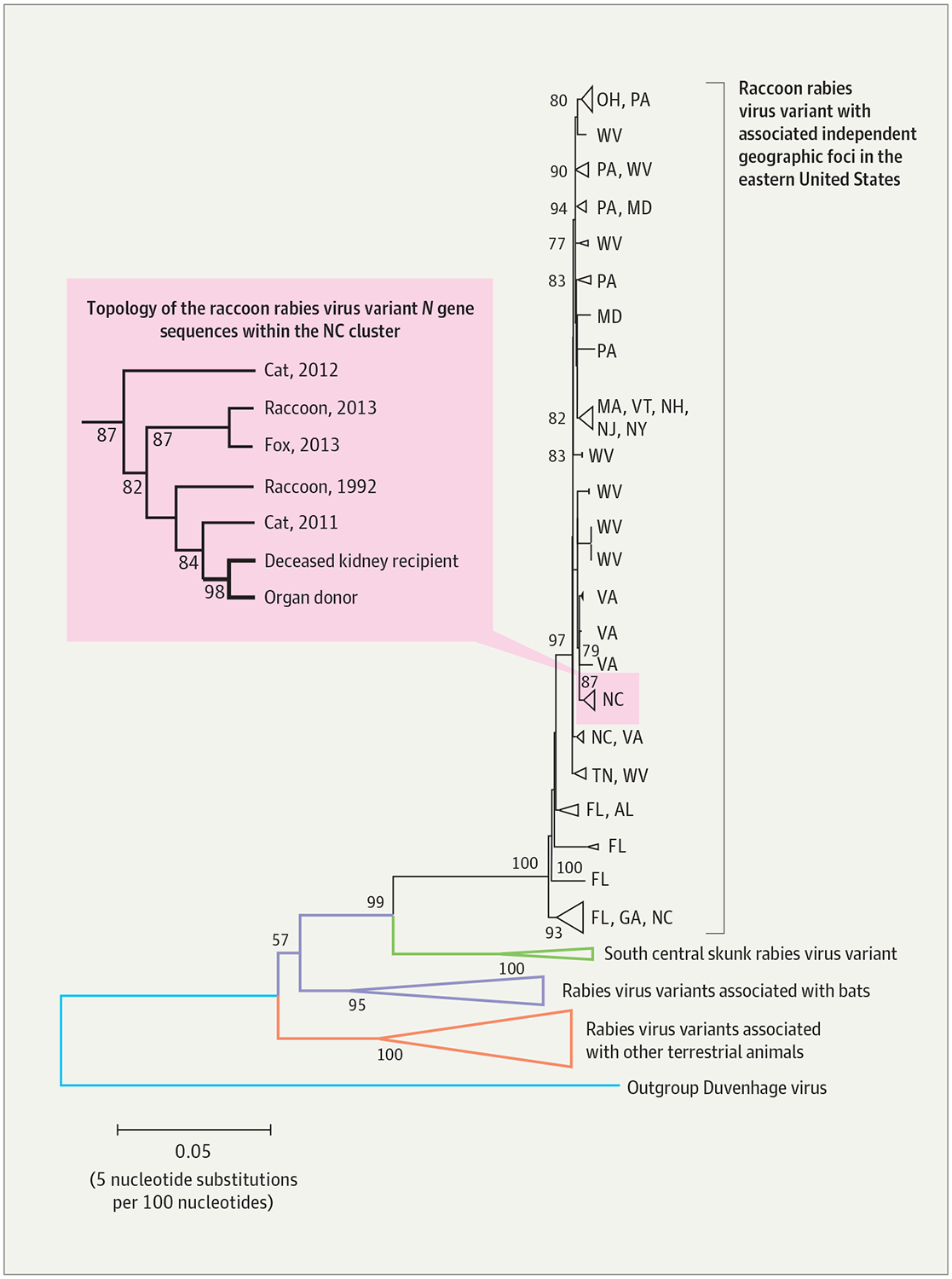
Phylogenetic Reconstruction of the Rabies Viruses Infecting the Deceased Kidney Recipient and Donor
The N gene sequences of the rabies viruses infecting the deceased kidney recipient and donor are shown in relation to N gene sequences of common rabies virus variants circulating in the United States. The phylogenetic reconstruction was generated with the neighbor-joining method under the maximum composite likelihood model to estimate nucleotide substitutions. The bootstrap method with 1000 iterations was used to assess the confidence of the branching pattern (numbers at the nodes). Branches are color coded according to reservoir host, with triangles representing collapsed branches with a common origin. The Duvenhage virus was used as an outgroup to better visualize the evolutionary relationships among rabies virus clades. Independent geographic foci associated with the raccoon rabies virus variant circulating in the eastern United States, in black, are shown along with the corresponding state (2-letter abbreviations). Enclosed in the rectangle on the left is a magnified projection showing the topology of the collapsed North Carolina cluster, which includes sequences obtained from various rabid animals, the deceased kidney recipient, and the donor.
