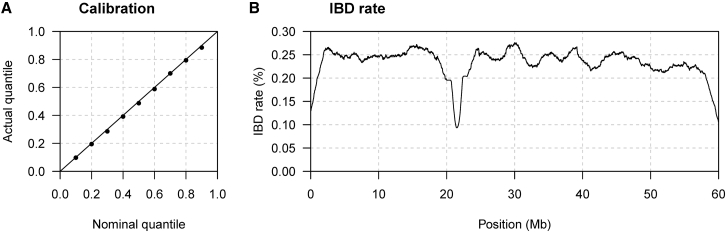
Figure 2.
Method Performance with Uneven Marker Density
Sequence data on 2,000 individuals were simulated under a constant effective population size. Markers located between 20 and 23 Mb were removed, and marker density varies every 100 kb (see Material and Methods). The true haplotype phase is used in the analysis.
(A) Quantile-quantile plot assessing the calibration of the estimated endpoint uncertainty. The actual quantile (y axis) corresponding to a given nominal quantile (x axis) is the proportion of segments for which the reported nominal quantile of the right endpoint is greater than the true right endpoint (points on the plot). The line is shown for comparison. Results for the left endpoints are similar but are not shown.
(B) The y axis is the IBD rate, which is the percentage of pairs of haplotypes for which the position on the chromosome is covered by an estimated IBD segment with length >2 cM for the haplotype pair. Estimated IBD segment endpoints are the posterior medians. The IBD rate is calculated at 10 kb intervals.