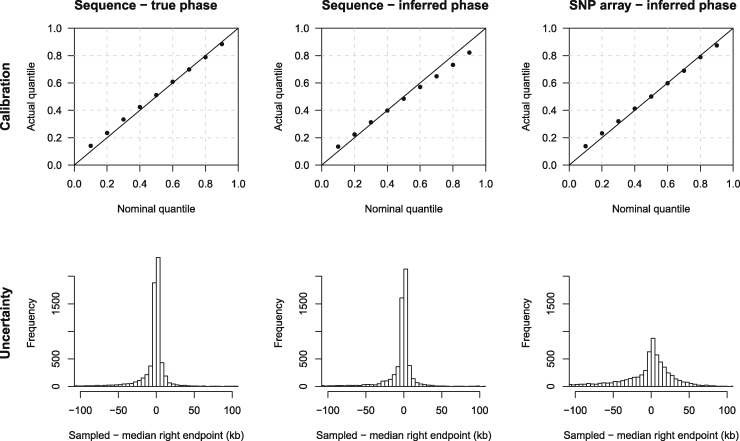
Figure 3.
Method Performance on UK-like Simulated Sequence and SNP Array Data
The data comprise 50,000 individuals simulated from a UK-like demographic history (see Material and Methods), with a genotype error rate of 0.02%. True IBD segment endpoints were determined for 1,000 individuals, and these individuals were used to generate the results in this figure. The top row shows quantile-quantile plots that assess the calibration of the estimated endpoint uncertainty. The line is shown for comparison. The actual quantile (y axis) corresponding to a given nominal quantile (x axis) is the proportion of segments for which the reported nominal quantile of the right endpoint is greater than the true right endpoint. The bottom row shows histograms of the right endpoint sampled from the estimated posterior distribution minus the posterior median right endpoint. The histograms represent the distribution of uncertainty, averaged over segments. Histogram bin widths are 5 kb. Results for the left endpoints are similar but are not shown. The left column is for analysis using the true haplotype phase. The middle column is for analysis using haplotype phase inferred using Beagle 5.1. The right column is for data thinned to match a SNP array with 500,000 markers genome-wide (10,000 markers in the simulated 60 Mb interval), and with haplotype phase inferred using Beagle 5.1.