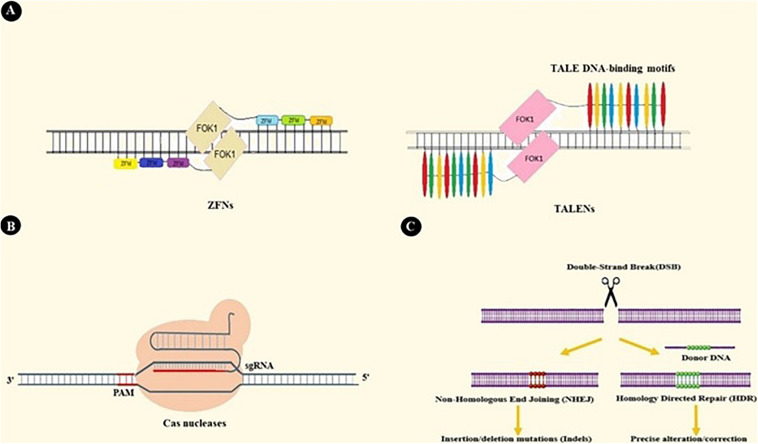
FIGURE 2.
ZFNs, TALENs, and CRISPR-based genome editing. (A) ZFNs and TALENs are nucleases that operate based on protein-DNA interactions. Assembling Zinc Finger Motifs (ZFMs) and TALEs into larger complexes increases specificity. (B) CRISPR/Cas9 binds to DNA under the guidance of single guide RNA (sgRNA). sgRNA is a chimeric RNA constructed by fusing crRNA and tracrRNA to simplify the guidance system. As a result, Cas9-sgRNA is the most extensively used system in CRISPR based applications (23). For DNA recognition, many CRISPR systems also need Protospacer Adjacent Motif sequence (PAM) adjacent to the crRNA target site. PAM sequences are specific to each type of nuclease (e.g., NGG sequence is specific to SpCas) (21, 198). (C) Double-Stranded Breaks (DSBs) created by Cas9 in DNA structure activate two DNA repair pathways: Non-Homologous End Joining (NHEJ) and Homology-Directed Repair pathways (HDR). NHEJ results in random insertions or deletions at the target site, so it involves knocking out genes in CRISPR-based applications. HDR is a precise pathway that repairs target DNA breakage by using a homologous donor DNA. This pathway takes part in techniques that need more precise genome editing, like insertion or deletion of the desired DNA fragment (199, 200).