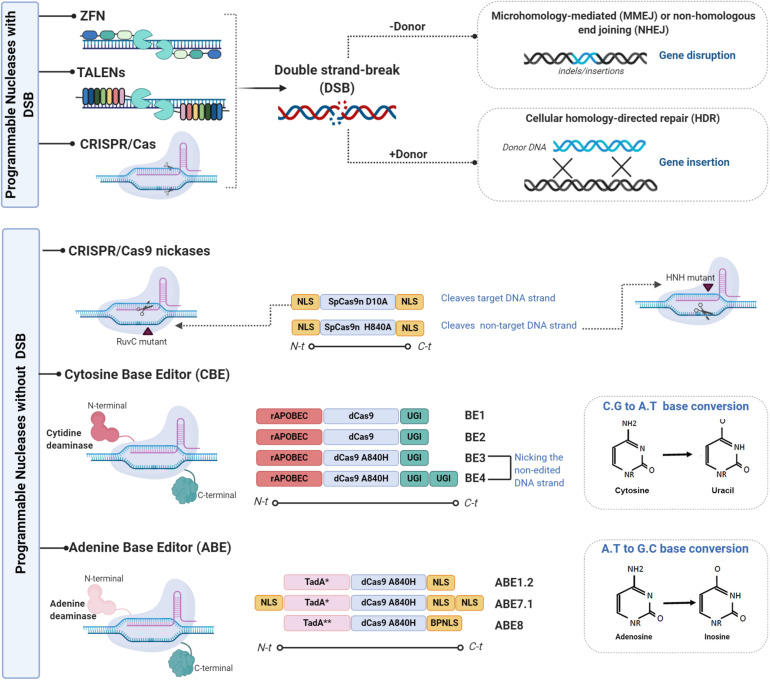
FIGURE 1.
Representative scheme of the different genome editing tools used to improve the immune system. There are two variants of genome editing technologies: those that introduce double-stranded breaks (DSBs) into DNA and those that enable genome editing without DSBs. The first variant is mainly composed of the components ZFNs, TALEN, and CRISPR/CAS which are used to enhance immune system capacity. ZFNs are chimeric proteins containing a DNA binding domain (3–5 zinc-finger domains) and a Fok1 endonuclease domain (114). Each zinc-finger domain is specifically designed to bind to virtually any DNA sequence. ZFN cleavage activity needs to be dimerized given that Fok1 acts as a dimer. Two ZFNs therefore need to be designed, each targeting a DNA sequence separated by a short sequence from the recognition site of the other ZFNs in a head-to-head fashion. As ZFNs, TALENs contain two different domains: the DNA binding domain of the TALE protein designed to bind the desired sequence (10) and the Fok1 endonuclease domain (11). As TALENs only act as dimers, two TALENs must be designed to bind to the target locus in a face-to-face fashion to cleave the target sequence (110). CRISPR/Cas, the last described SENs, is the easiest to design and the most versatile gene editing tool. It is derived from the adaptive immune system of prokaryotes, which provides a defense mechanism against certain viral infections and plasmids. The CRISPR/Cas protein forms a complex with the RNA molecules crRNA (CRISPR RNA) and crRNA (tracrRNA) which guides the Cas protein to the target DNA and produces the cleavage (12). The only part that needs to be changed to specifically cut a new target site in the genome is 20 crRNA nucleotides (13, 14). Various modifications to the gRNA design and to the Cas9 protein have been made to reduce off-target activity. A mutated Cas9 nickase has been generated to expand the CRISPR genome editing system (111). The second group is mainly composed of CRISPR-CAS nickases variants and of two base editor (BE) variants, the cytosine base editor (CBE) and the adenine base editor (ABE). The first variant is based on simple APOBEC deaminase system named BE1, which fuses APOBEC1 and dead Cas9 from Streptococcus pyogenes with D10A and H840A mutations (28). Its lower efficiency is attributable to uracil DNA glycosylase (UDG) which catalyzes the removal of U from DNA in cells and initiates base-excision repair (BER), thus converting the U:G pair to the C:G pair (115). The uracil-DNA glycosylase inhibitor (UGI) was fused to the C terminus of BE1 to create the second-generation BE2 system, with an improved base editing yield of 50%. A further improvement was implemented for the third-generation BE3 system. The improved BE4 base editor contains a rAPOBEC cas9 linker expanded to 32 amino acids, a Cas9n-UGI linker expanded to 9 amino acids, as well as the addition of a second copy of UGI to the C terminus of the constructs (116); BE3 and BE4 have been validated for use as base editors of human primary T cells (55). The replacement of the APOBEC1 component in BE3 with natural adenine deaminase Escherichia coli TedA led to the creation of the first adenine base editor ABE which was followed by ABE1.2 After several target mutations and optimizations, the ABE7.1 base editor was released. This was followed by the latest version ABE8 with its base editing facility particularly for HSPCs and human primary T cells (33). Figures were created by BioRender.com.