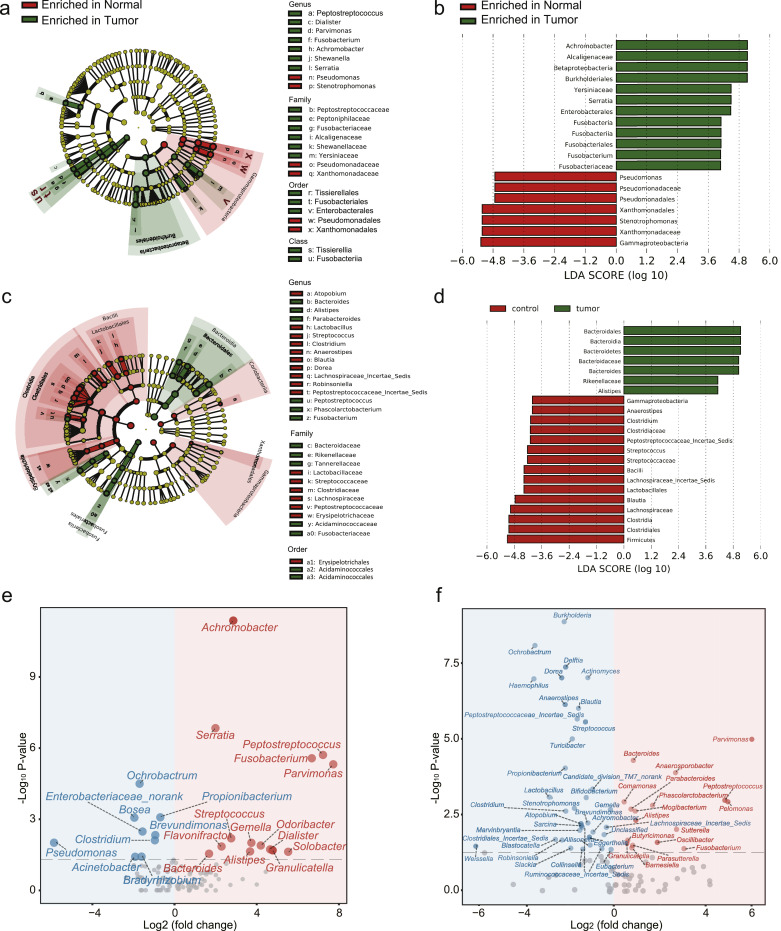
Fig. 1.
16S rRNA sequencing analysis of the gut microbiome in Cohort 1 (mucosal) and Cohort 2 (fecal). (a) A cladogram representation of data in Cohort 1. (Green) Tumour-enriched taxa; (red) taxa enriched in normal tissue. The brightness of each dot is proportional to its effect size. (b) Linear discriminant analysis (LDA) coupled with effect size measurements identifies the significant abundance of data in (a). Tumour-enriched taxa are indicated with a positive LDA score (green), and taxa enriched in normal tissue have a negative score (red). Only taxa meeting an LDA significant threshold of 4 are shown. (c) A cladogram representation of data in Cohort 2. (Green) Tumour-enriched taxa; (red) taxa enriched in normal tissue. The brightness of each dot is proportional to its effect size. (d) LDA coupled with effect size measurements identifies the significant abundance of data in (c). Tumour-enriched taxa are indicated with a positive LDA score (green), and taxa enriched in normal tissue have a negative score (red). Only taxa meeting an LDA significant threshold of 4 are shown. (e) Volcano plot of significantly (Wilcoxon test, P < 0.05) altered genera between normal and CRC samples in Cohort 1. Tumour-enriched genera are colored in red and genera enriched in normal samples are colored in blue. (f) Volcano plot of significantly (Wilcoxon test, P < 0.05) altered genera between normal and CRC samples in Cohort 2. Tumour-enriched genera are colored in red and genera enriched in normal samples are colored in blue.