Fig 1.
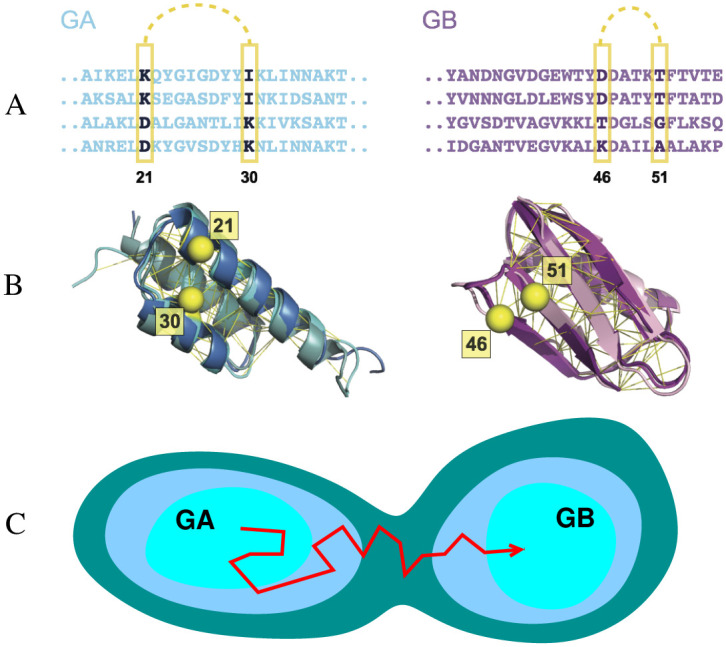
Sequence-based models for the GA and GB domains of streptococcal protein G. Many sequences (A) fold to each structure (B): e.g. structures of three naturally occurring sequences with the GA fold (pdb ID 2fs1, 1gjs and 2j5y) and three with the GB fold (pdb ID 1pga, 2lum and 1igd) are shown on the left and right respectively. Contacts between pairs of residues in the native structure (Cβ atoms of example pairs in yellow) impose mutual constraints on the types of residues which can occupy these positions in the sequence alignment. For instance, strong covariance is detected between the amino acids at residue 21 and 30 for GA sequences and between residues 46 and 51 for GB sequences. The Cβ atoms of these residues are illustrated in yellow sphere. The UniProtKB ID of these example sequences for GA are Q51918_FINMA, G5KGV3_9STRE, G5K7M6_9STRE and Q56192_STAXY. And the ones for GB are SPG1_STRSG, E4KPW8_9LACT, F9P4J6_STRCV and G5JZF8_9STRE. (C) Simple model for the emergence of new folds via evolutionary drift in sequence space between basins of attraction corresponding to the GA and GB domains.
