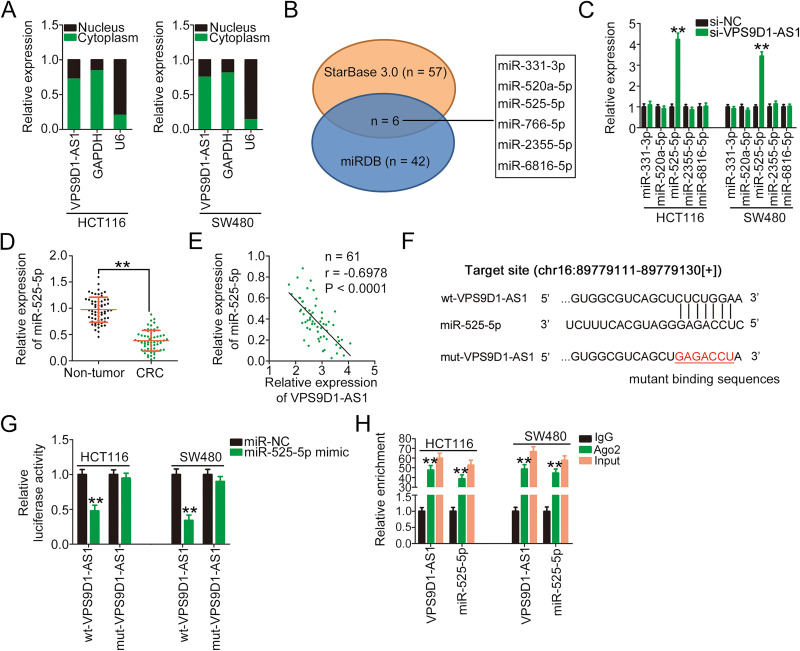
Figure 2.
VPS9D1-AS1 functions as a miR-525-5p sponge in CRC cells. (A) The isolation of cytoplasmic/nuclear HCT116 and SW480 cell fractions was performed, followed by qRT-PCR to determine the subcellular location of VPS9D1-AS1. (B) Schematic illustration of the putative miRNAs interacting with VPS9D1-AS1. (C) qRT-PCR was performed to determine the relative expression levels of miR-331-3p, miR-520a-5p, miR-525-5p, miR-2355-5p, and miR-6816-5p in HCT116 and SW480 cells after VPS9D1-AS1 downregulation. (D) qRT-PCR was performed to determine miR-525-5p expression in 61 pairs of CRC tissues and adjacent non-tumor tissues. (E) Pearson’s correlation analysis showed an inverse correlation between VPS9D1-AS1 and miR-525-5p expression in the 61 CRC tissues. (F) The wild-type and mutant binding sites of miR-525-5p in VPS9D1-AS1 were presented. (G) Luciferase activity of HCT116 and SW480 cells transfected with wt-VPS9D1-AS1 or mut-VPS9D1-AS1 and miR-525-5p mimic or miR-NC. (H) RIP assays followed by qRT-PCR analyses were conducted to test miR-525-5p and VPS9D1-AS1 enrichment in HCT116 and SW480 cells associated with Ago2. **P < 0.01.
Abbreviations: GAPDH, glyceraldehyde 3-phosphate dehydrogenase; U6, U6 small nuclear RNA; VPS9D1-AS1, VPS9D1 antisense RNA 1; si-VPS9D1-AS1, small interfering RNA targeting VPS9D1-AS1; si-NC, negative control small interfering RNA; miR-525-5p, microRNA-525-5p; wt, wild-type; mut, mutant; miR-NC, miRNA mimic negative control; Ago2, Argonaute2; CRC, colorectal cancer.