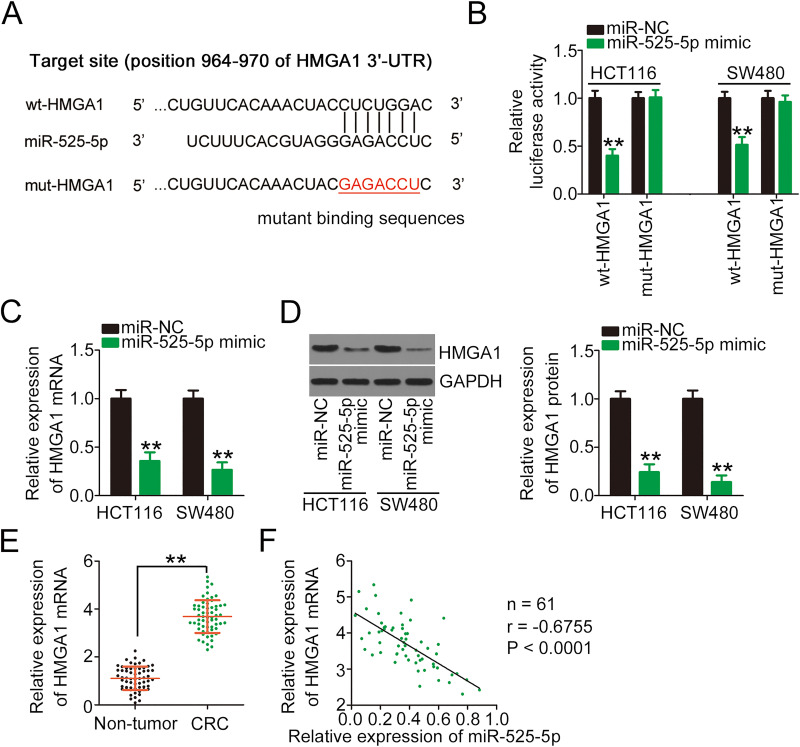
Figure 4.
HMGA1 is a direct target of miR-525-5p in CRC cells. (A) Bioinformatics analysis predicted that the 3′-UTR of HMGA1 contains a miR-525-5p binding site; the mutated binding sequences were also shown. (B) Luciferase activity was determined in HCT116 and SW480 cells cotransfected with wt-HMGA1 or mut-HMGA1 and miR-525-5p mimic or miR-NC. (C, D) qRT-PCR and Western blotting were performed to determine the mRNA and protein levels of HMGA1 in HCT116 and SW480 cells in the presence of miR-525-5p mimic or miR-NC by. (E) qRT-PCR was performed to determine the mRNA level of HMGA1 in 61 pairs of CRC tissues and adjacent non-tumor tissues. (F) Pearson’s correlation analysis of experimental data presented an inverse correlation between the expression levels of miR-525-5p and HMGA1. **P < 0.01.
Abbreviations: HMGA1, high-mobility group AT-hook 1; wt, wild-type; mut, mutant; miR-NC, miRNA mimic negative control; miR-525-5p, microRNA-525-5p; 3ʹ-UTR, 3′-untranslated region; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; CRC, colorectal cancer.