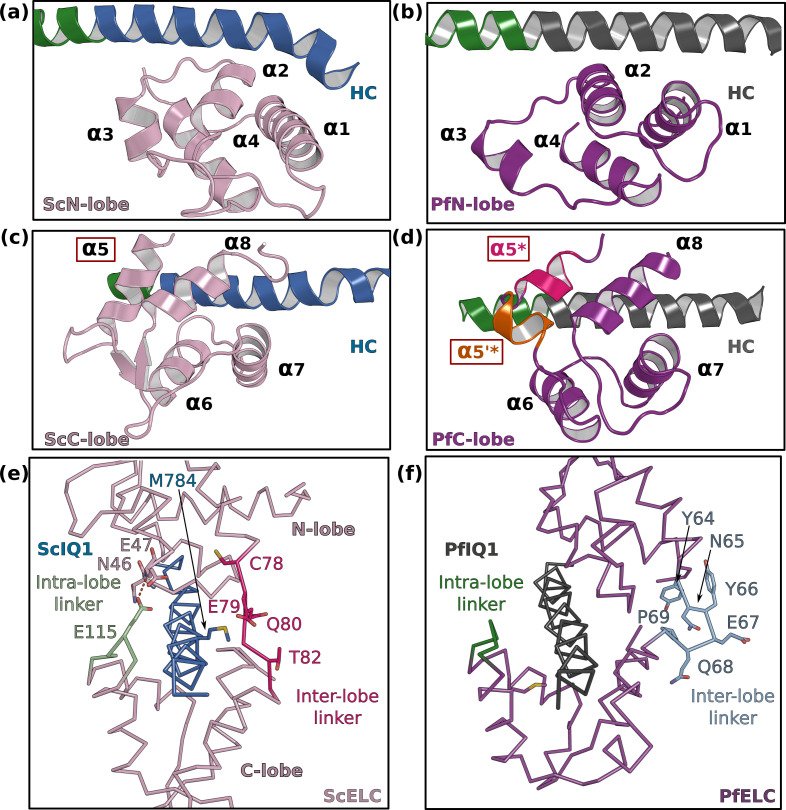
Figure 3. PfELC adopts an atypical fold.
(a) The N-lobe of scallop myosin 2 ELC (ScMyo2) is compared to (b) the N-lobe of PfELC. The two lobes are conserved in structure, and adopt a closed conformation. In contrast, the C-lobe of ScMyo2 ELC (c) and PfELC (d) differ. The most divergent feature is the much shorter α5* helix (comprised of only one turn) and the presence of the short α5’* helix in PfMyoA. (e) shows the elongated inter-lobe linker of ScELC (red) compared with (f) the kinked, hairpin-like inter-lobe linker of PfELC (light blue). The specific structure of the inter-lobe linker makes the PfELC structure more compact than canonical ELCs and no direct interaction can occur between the lobes in PfMyoA to encircle the heavy chain, contrary to what is seen in the case of ScMyo2.