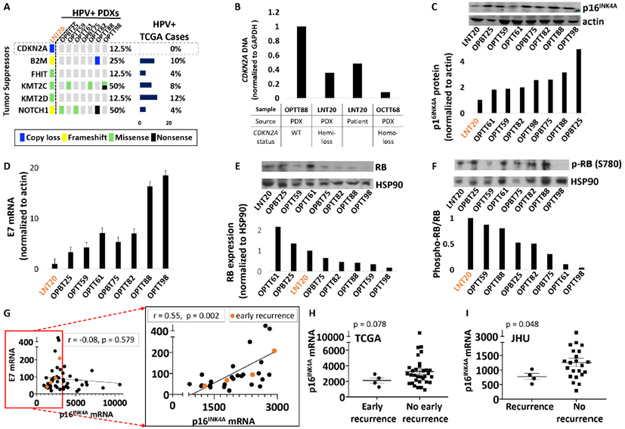
Figure 5. Reduced p16INK4A levels in an atypical HPV+ PDX are shared by recurrent HPV+ cases.
A. Tumor suppressors mutated or deleted in LNT20 relative to the other HPV+ PDXs and TCGA cases. B. CDKN2A DNA levels in the LNT20 PDX and its tumor of origin, relative to control PDXs with WT or deleted CDKN2A. C. p16INK4A protein by western blot (top) with band densities normalized to actin (bottom) in HPV+ PDXs. D. HPV16 E7 mRNA levels normalized to actin in HPV+ PDXs. E RB protein (top) with corresponding band densities normalized to HSP90 (bottom) in HPV+ PDXs. F. phospho-RB protein (Ser780) (top) and phospho-RB to total RB ratios (bottom). G. E7 vs. p16INK4A mRNA in all TCGA HPV+ HNSCCs (left) and those with a normalized p16INK4A mRNA level of <3000 (right inset). Recurrences within 2 years (early recurrence) are highlighted in orange. Pearson correlation coefficients and p-values based on two-tailed Student t-distribution are shown for each plot. H. p16INK4A mRNA expression in TCGA HPV+ HNSCCs that recurred within 2 years (early recurrence) or were disease-free at the time of last early recurrence. Bars represent mean ± SEM. p-values were determined by randomization test (n = 100,000 permutations) with Welch’s t-statistic. I. p16INK4A expression in recurrent vs. non-recurrent HPV+ cases with matched follow-up duration in the JHU cohort. Bars represent mean ± SEM. p-values were determined by randomization test (n = 100,000 permutations) with Welch’s t-statistic.