Fig. 3.
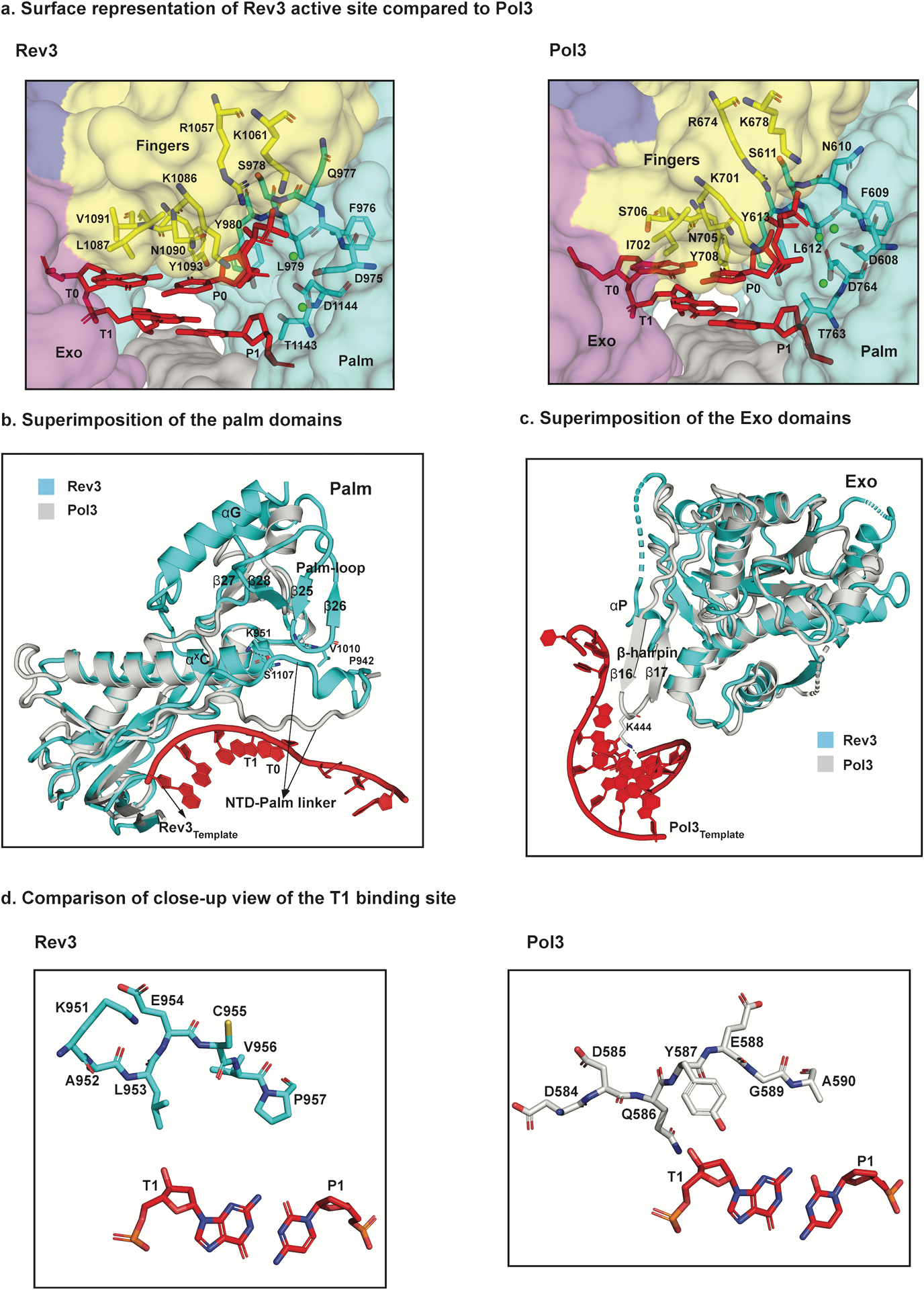
Structural basis for fidelity and mismatch extension. a, Surface representation of a close-up view of the active sites of Rev3 and Pol3 depicting conserved residues interacting with the incoming nucleotide (dCTP) as well as the templating base (G). b, Superimposition of the palm domain of Rev3 and Pol3. The Rev3 template DNA strand is shown in red and the T1 base is highlighted. In comparison to Pol3, the palm-loop (β25 and β26) is a unique Rev3 structural element that interacts with the NTD-palm linker. Key residues in the NTD-palm linker are shown as sticks interacting with the palm-loop residues as well as the αXC palm helix. Another unique structural element, αG, which interacts with β27 and β28 is also highlighted. c, Superimposition of the exonuclease domains of Rev3 and Pol3. Overlay of the exonuclease domains of Rev3 and Pol3 shows a much shorter and disordered helical loop in Rev3 in comparison to a well-defined β-hairpin in Pol3. The Pol3 DNA is highlighted in red. d, Comparison of the T1 binding site in Rev3 and Pol3, showing details of the interaction between the NTD-palm linker region and the T1 base. The residue E954 in Rev3 points away from the T1 base, whereas residue Y587 stacks against the sugar in Pol3. L953 in Rev3 is the only residue close to the T1 base, resulting in a less constrained pocket in comparison to Pol3.
