Extended Data Fig. 2. Cryo-EM data collection and processing of Polζ-DNA-dCTP complex.
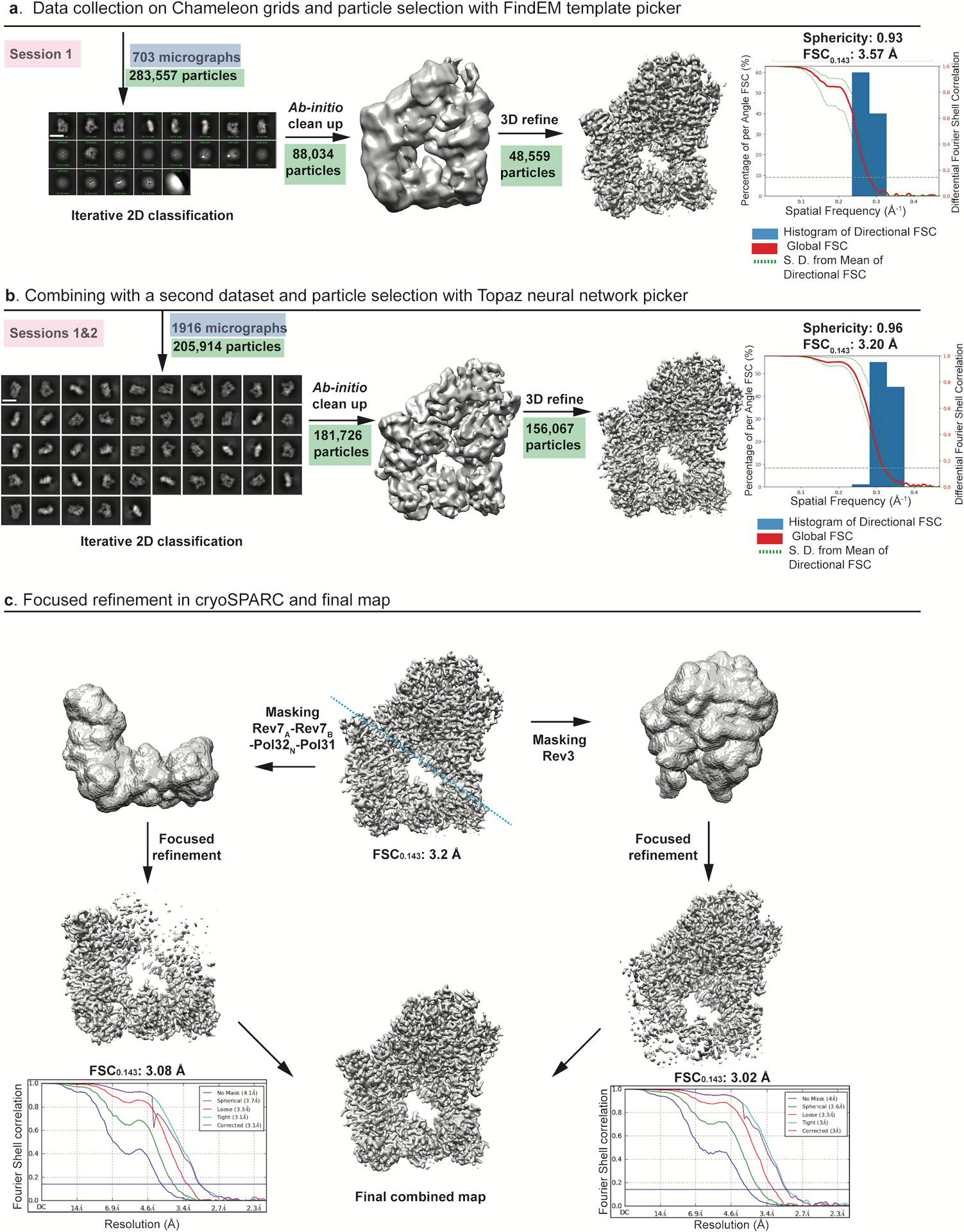
a, Data were collected on Chameleon grids and particles from one session were picked with template based picker (FindEM) and processed in cryoSPARC to give a consensus map with a FSC0.143 of 3.57 Å. Major stages of processing are shown schematically and particles involved at each stage are highlighted in green. Scale bar = 137 Å. b, Final particles from two sessions were merged and used to train Topaz. Data processing from Topaz picked particles in cryoSPARC2 improved the sphericity. A schematic representation of the improved consensus map displaying a FSC0.143 of 3.2 Å is shown. Scale bar = 137 Å. c, Focused refinement of the final volume was done in cryoSPARC2. Masks were created (along the blue dashed line) for 3D refinement of Rev3 and accessory subunits separately to give consensus maps at 3.02 Å and 3.08 Å, respectively..
